Preparation method of developing Phaseolus vulgaris SSR primers by magnetic bead enrichment
A technology of kidney beans and magnetic beads, applied in the field of bioengineering
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
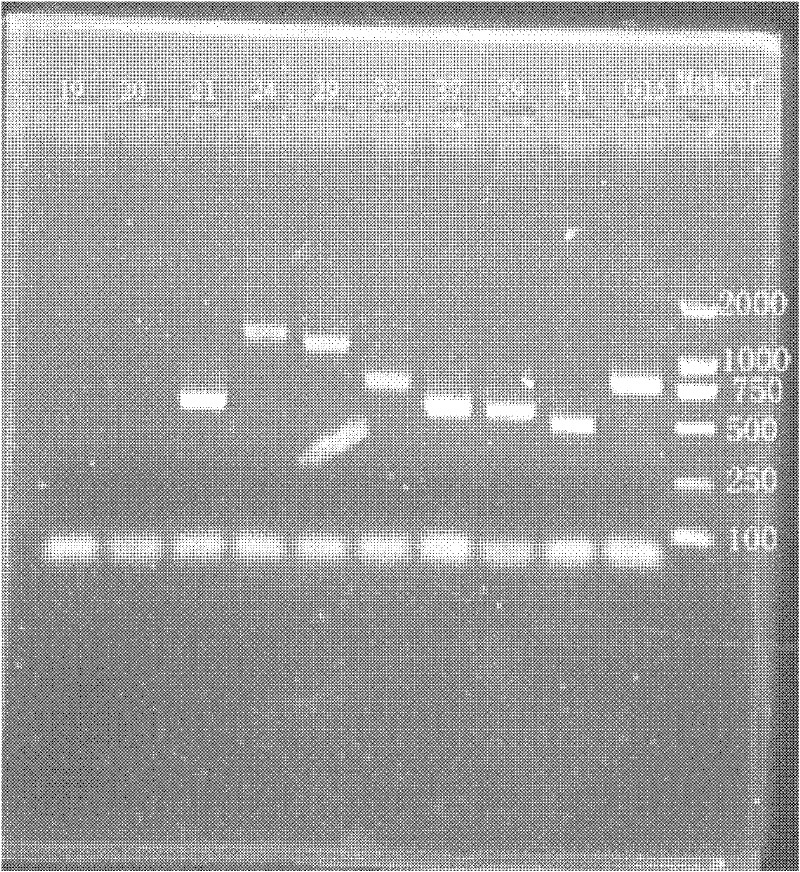
Image
Examples
Embodiment 1
[0060] A kind of magnetic bead enrichment method develops the preparation method of kidney bean SSR primer, and its steps are:
[0061] A. Extract the genome DNA of kidney bean, carry out enzyme digestion to the genome, and establish the AFLP-PCR library:
[0062] The bean variety Qiao Yu (an excellent bean variety provided by Wuhan Institute of Vegetable Science, creeping, green leaves, light yellow flowers). This variety is a premium string bean variety with the general characteristics of string bean varieties. Plant according to conventional cultivation management and fertilizer and water measures, wait for the seedlings to grow 5-6 leaves, take 0.4 g of young leaves and use the CTAB method to extract genomic DNA.
[0063] (1) Add 4ml of 3×CTAB extract (containing 0.2% β-mercaptoethanol (W / V)) to a 10ml Eppeddorf tube and preheat in a 65°C water bath.
[0064] (2) Take 0.4 g of young leaves of kidney bean and put them in a mortar, add appropriate amount of liquid nitrogen...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com