High flux detection method and biochip based on nucleic acid address coding
An address coding and detection method technology, which is applied in the field of high-throughput detection methods and biochips based on nucleic acid address coding, can solve the problems of lack of protein detection method biochips and limit the progress of proteomics research, and achieve reliable detection results. Improve detection types and good stability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
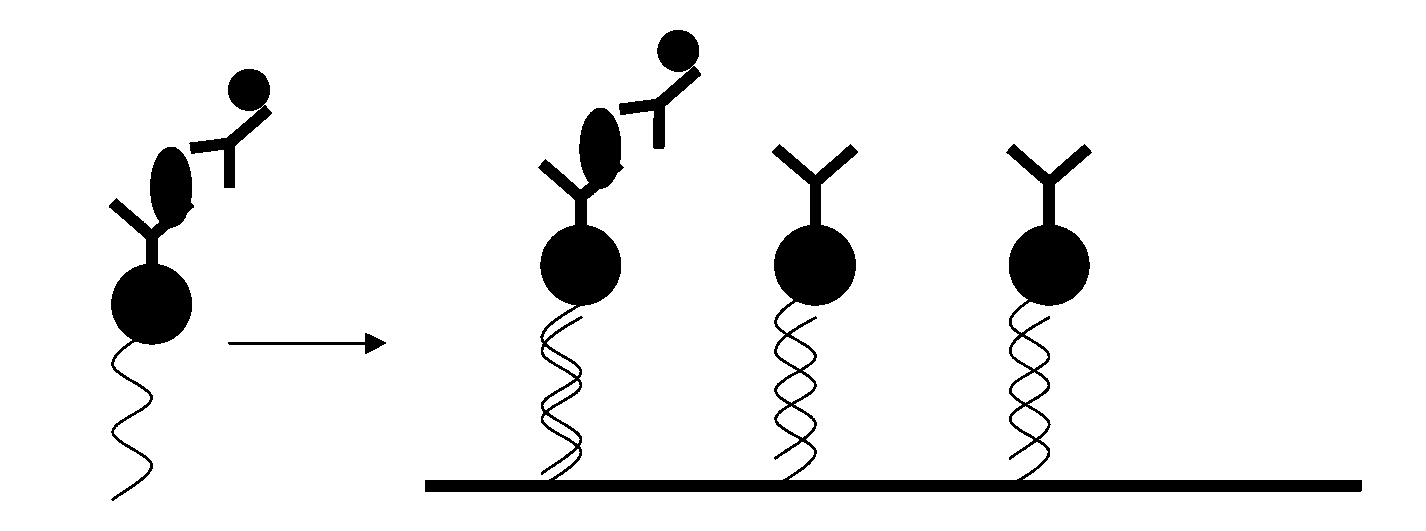
[0029] A high-throughput detection method based on nucleic acid address coding, comprising the following steps:
[0030] 1) immobilizing the first single-stranded nucleic acid array on a solid phase carrier;
[0031] 2) coupling a detection substance capable of specifically reacting with the analyte to a second single-stranded nucleic acid to obtain a detection body, wherein the second single-stranded nucleic acid is complementary to the first single-stranded nucleic acid;
[0032] 3) Mix and react the analyte with the analyte, then add the labeled secondary antibody, and continue the reaction to obtain the analyte-analyte-secondary antibody complex; combine the above complex with the first single-chain immobilized on the solid phase carrier For nucleic acid hybridization, wash to remove unhybridized reactants; or mix and react the test substance with the test substance, hybridize with the first single-stranded nucleic acid immobilized on the solid phase carrier, and wash; the...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com