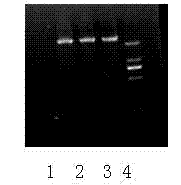
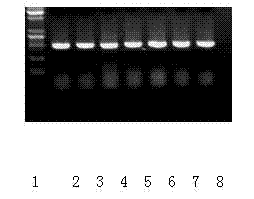
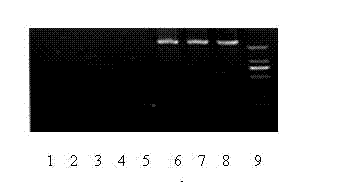
Method for identifying tomato yellow leaf curl virus resistance
A technology for tomato yellowing leaf curl and virus, which is applied in the directions of biochemical equipment and methods, horticultural methods, and botanical equipment and methods, and can solve problems such as inability to identify TYLCV resistance.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0108] 1. Extraction of plant total DNAD
[0109] (1) Take the leaves 30 days after inoculation, add liquid nitrogen to grind them into powder, and quickly transfer them into 1.5 ml Eppendorf tubes;
[0110] (2) Add 800 μl of CTAB extraction buffer, mix well (CTAB is preheated in a water bath at 65°C), oscillate gently every 5 minutes, centrifuge at 12000 r / min for 20 minutes, and centrifuge for 15 minutes;
[0111] (3) Carefully draw the supernatant, add an equal volume of phenol:chloroform (400 μl each) solution, mix well, centrifuge at 12000 r / min at 4 °C for 10 min;
[0112] (4) Carefully draw the supernatant, add an equal volume of chloroform, mix well, centrifuge at 12000 r / min at 4°C for 10 min.
[0113] (5) Repeat step (4) 1-2 times until the protein layer does not appear;
[0114] (6) Take the supernatant, precipitate at -20°C for 1 hour, centrifuge at 12000 r / min at 4°C for 10 minutes;
[0115] (7) Discard the supernatant and wash the precipitate twice with 70% et...
Embodiment 2
[0167] Construction of Infectious Vector of TYLCV-TJ Isolated from Tianjin Area
[0168] The 1.4A-pMD plasmid containing 1.4A TYLCV and the plant expression vector pRI 101-An were digested with KnpI and SalI to recover the target fragment. Then use T4 ligase to connect the target fragment with the expression vector to obtain 1.4A-pRI. The ligation product was transformed into E. coli DH5α.
[0169] Digestion:
[0170] Take a 0.5 mL eppendorf tube and add the following reagents in sequence
[0171] DNA 200ng
[0172] Restriction enzyme 0.5 μL
[0173] 10× Enzyme Reaction Buffer 2 μL
[0174] Add dd HO to 20 μL
[0175] The results were checked by agarose gel electrophoresis after enzyme digestion and digestion at 37°C for 1 hour.
[0176] For connection and transformation, see 2.3
Embodiment 3
[0178] Transformation of recombinant vectors to Agrobacterium:
[0179] Preparation of Competent Cells of Agrobacterium LBA4404
[0180] 1) Agrobacterium activation: Line the Agrobacterium LBA4404 stored at -70°C on YEB solid medium containing kanamycin (50 ug / mL) and rifampicin (50 ug / mL), and place in a 28°C incubator Inverted culture in the medium for about 48h until the colony grows;
[0181] 2) Pick a single colony and inoculate it in 3 mL of YEB liquid medium containing kanamycin (50 ug / mL) and rifampicin (50 ug / mL), shake at 28°C at 220 rpm / min until OD600≈0.5;
[0182] 3) Take 1.0 mL of the bacterial solution in a 1.5 mL sterile centrifuge tube and place on ice for 10 min;
[0183] 4) Take out the centrifuge tube from ice, centrifuge at 5,000 rpm / min for 30 s in a desktop centrifuge, and discard the supernatant;
[0184] 5) Suspend the precipitate fully with 1.5 mL 0.5 mol / L NaCl, do not shake vigorously, and then place it on ice for 20 min;
[0185] 6) Centrifuge ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com