Method for ultrahigh-level and targeting expression of salmon calcitonin protein in oleosin of transgenic rape by using chromosome substitution technology
A salmon calcitonin and gene technology, which is applied in the field of specific expression technology with vegetable oil bodies, can solve the problems of low probability of homologous recombination and difficulty in transgenic plants
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0042] Embodiment 1, the cloning of Saccharomyces cerevisiae RAD54 gene
[0043] 1. Extraction of Saccharomyces cerevisiae genome
[0044] 1) Pick a single colony of yeast (purchased from China Industrial Microbiology Culture Collection Management Center, No. 31141), inoculate in 10ml YPD (1% yeast extract, 2% glucose, 1.5% agar powder, 2% peptone) liquid medium Incubate overnight in a shaker at 30°C;
[0045] 2) Take 1ml of the yeast liquid cultured overnight, centrifuge at 5000rpm for 5min, remove the supernatant, add 1ml of sterile water, resuspend the pellet, centrifuge at 5000rpm for 1min, remove the supernatant, add 200μl of bacteria-breaking buffer (1% SDS, 2 % TritionX-100, 100mM NaCl, 10mM Tris-Cl pH8.0, 1mM EDTA), shake vigorously.
[0046] 3) Add 200 μl phenol / chloroform / isoamyl alcohol (volume ratio 25:24:1), shake vigorously, centrifuge at 12,000 rpm for 5 minutes, take the supernatant, add an equal volume of chloroform, shake it upside down, centrifuge at 12,00...
Embodiment 2
[0055] Embodiment 2, the construction of RAD54 plant expression vector pRAD3301
[0056] 1. Construction of intermediate vector p3301
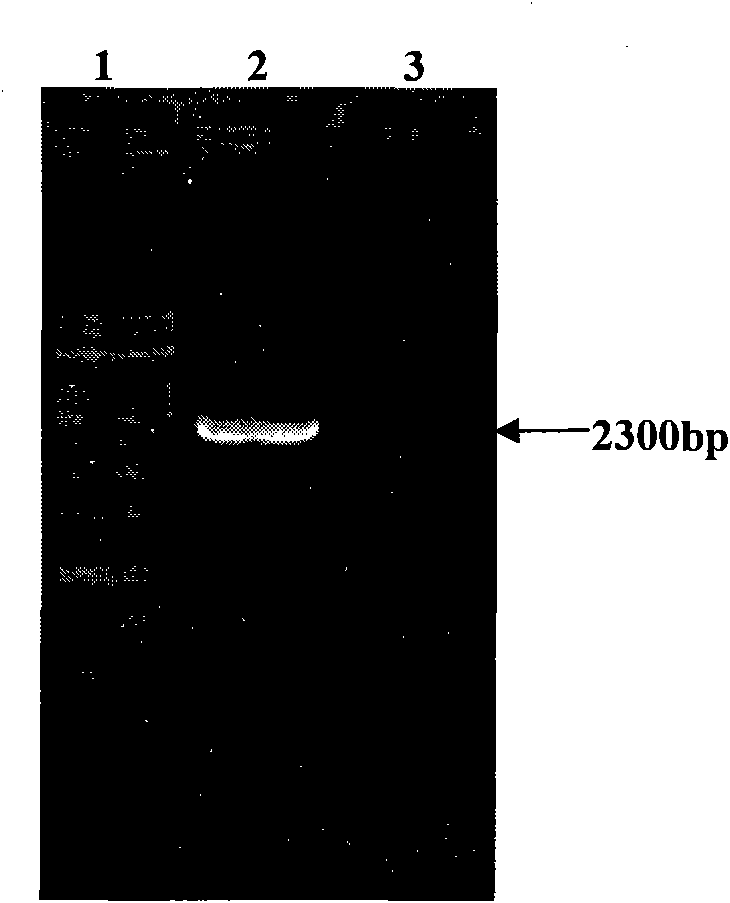
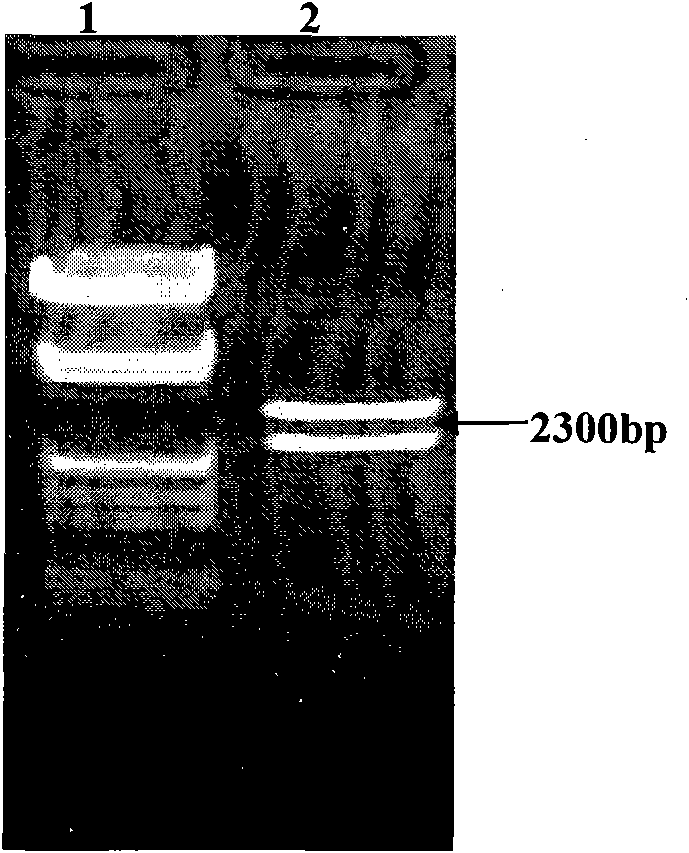
[0057] The expression cassette of gene pRAD3301 constructed in this example includes the following gene expression regulatory elements: 35S promoter, RAD54 gene and NOS terminator. First, pTΩ4A was double-digested with restriction endonucleases EcoRI and HindIII to recover a 999bp fragment, which was ligated with the same digested plant vector pCAMBIA3301 to obtain an intermediate vector named p3301. EcoRI and HindIII double enzyme digestion was used for verification, and the enzyme digestion identification results were as follows: image 3 As shown, the results prove that the pTΩ4A fragment has been linked to pCAMBIA3301.
[0058] 2. Construction of RAD54 plant expression vector pRAD3301
[0059] Plasmid RAD54 was digested with restriction endonuclease PstI, and a 2700bp fragment was recovered, which was ligated with vector p3301 cut with ...
Embodiment 3
[0060] Embodiment 3, the cloning of rapeseed oleosin gene 5' upstream sequence
[0061] Plant material: Rapeseed variety Zhongshuang No. 4, when seedlings are 4 weeks old.
[0062] 1. Design of chromosomal walking primers for the 5' upstream sequence of oleosin gene
[0063] The size of the homologous sequence is an important factor affecting the efficiency of gene targeting. When constructing a gene targeting vector, it is generally required to have a DNA homologous sequence of about 2 kb, but the 5' upstream sequence of the oleosin gene (sequence number M63985) registered on Genbank The size is only 944bp, for which, chromosome walking is needed to further clone the upstream sequence of oleosin.
[0064] According to the 5'UTR sequence of oleosin gene registered on Genbank and GenomeWalker TM According to the requirements of Universal Kit, design 2 specific primers, the primer sequences are as follows
[0065] GSP1: 5'-TAATCTCCGCCGCGTCTCTCTCTAAAC-3'
[0066] GSP2: 5'-CC...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com