Double carbonyl reductase mutant and its application
A technology of double carbonyl reductase and reductase, which is applied in oxidoreductase, application, genetic engineering, etc., can solve the problem of poor stereoselectivity and achieve high stereoselectivity and high conversion efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0047] Example 1: Cloning of double carbonyl reductase mutant gene
[0048] 1. PCR amplification of genes containing mutation sites
[0049] Using the principle of overlap extension PCR (OE-PCR), the double carbonyl reductase gene containing the mutation site was obtained through two-step PCR reactions.
[0050] Design a pair of upstream and downstream primers according to the known DNA sequence of double carbonyl reductase, where the underlined parts are the restriction sites of NdeI and BamHI respectively:
[0051] SEQ ID No.: 41 Primer NO.1: 5'-CAACAAGACATATGACCGGCATCACGAAT-3'
[0052] SEQ ID No.: 42 Primer NO.2: 5'-AATGGATCCTCAGTACCGGTAGAAGCCCT-3'
[0053] Design mutation primers containing mutation sites, as shown in the table below:
[0054] Table 1 Mutation primers of double carbonyl reductase
[0055]
[0056]
[0057] In the first step of PCR reaction, PCR amplification was performed using pET22b-DKR (see: NaturePrecedings, http: / / hdl.handle.net / 10101 / npre.2...
Embodiment 2
[0067] Example 2: Mutant Gene Sequencing and Sequence Alignment
[0068] The mutant monoclonal obtained by the method in Example 1 was sent to Shanghai Yingjun Biotechnology Co., Ltd. for sequencing, and the sequencing results were input into the BioXM2.6 software to compare the differences between the gene base sequence and the protein amino acid sequence before and after the mutation.
[0069] The results showed that the amino acid sequences of the 20 mutant proteins contained one or more mutation sites compared with the wild type. The specific mutant protein amino acid sequence and nucleotide sequence changes are shown in Table 2:
[0070] Table 2 Sequence changes of double carbonyl reductase mutants
[0071]
[0072]
[0073] .
Embodiment 3
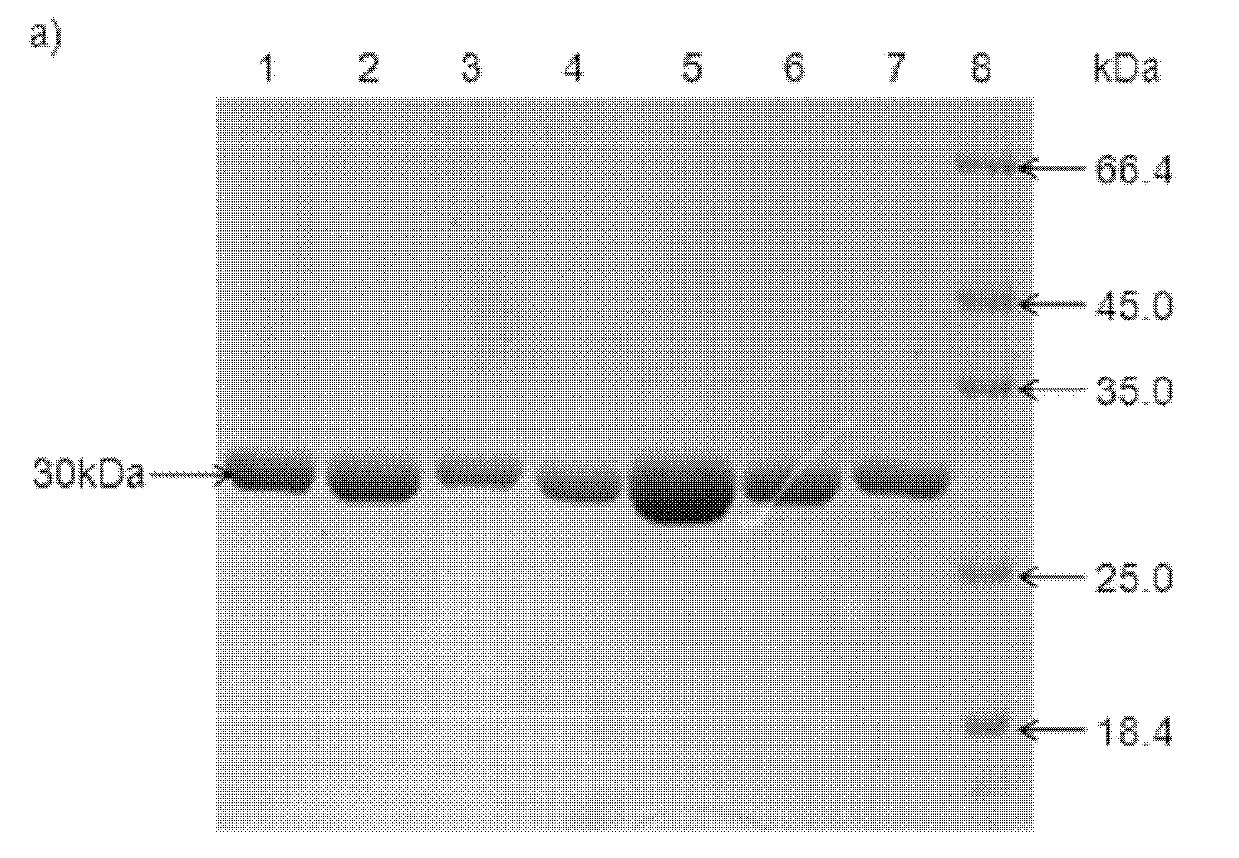
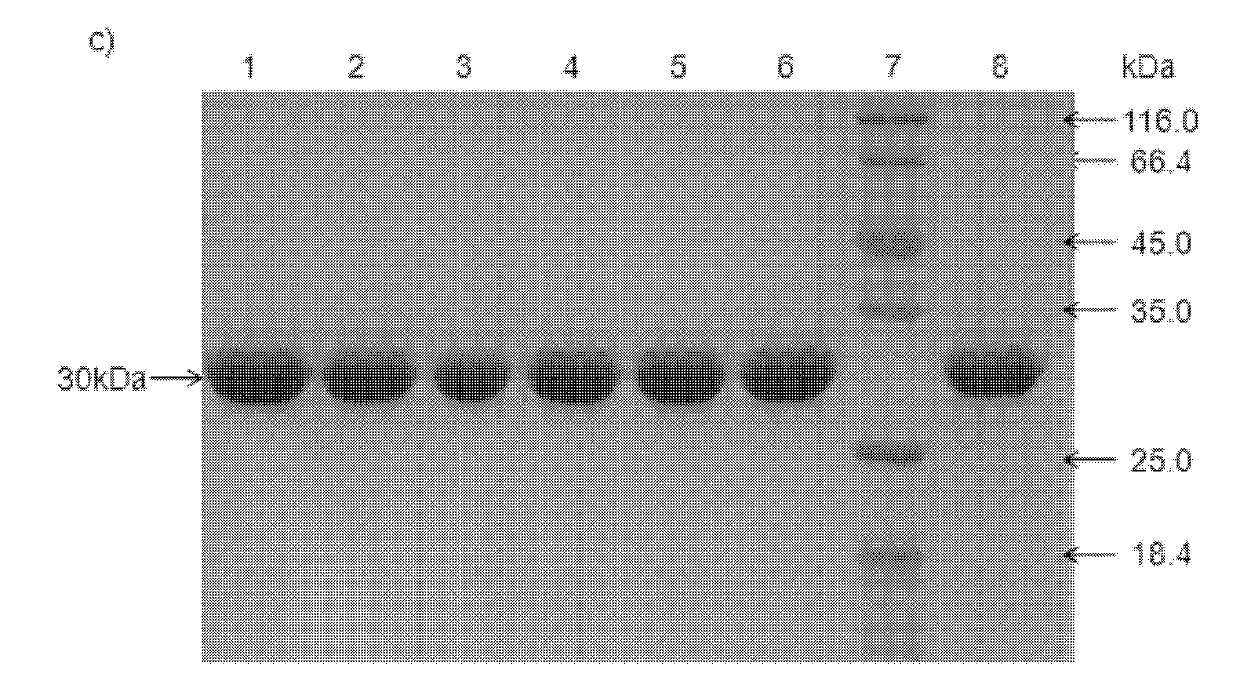
[0074] Example 3: Expression and purification of double carbonyl reductase mutants in Escherichia coli
[0075] 1. Expression of double carbonyl reductase mutant protein
[0076] The recombinant engineered bacteria obtained by the method described in Example 1 were inoculated into 50 ml of LB liquid medium containing 100 μg / ml ampicillin, and cultured at 37° C. for 12 to 16 hours. Then take 500μl and inoculate it into 50ml of fresh LB liquid medium containing ampicillin, shake and culture at 37℃ until OD 600nm When the value is about 0.8-1.0, add IPTG to a final concentration of 50 μM, and then continue to culture at 15° C. for 12-16 hours to induce expression. The bacterial cells were collected by centrifugation at 13,500 rpm / min, and washed once with 0.1M phosphate buffer solution of pH 7.0. Then the cells were disrupted with a high-pressure cell disruptor, centrifuged at 13,500 rpm / min for 30 min, and the supernatant was collected after centrifugation to obtain the crude ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com