Primers, probes, methods and kits for detecting Campylobacter coli
A technology for the detection of Campylobacter coli, which is applied in the field of molecular biology, can solve the problems of harsh culture conditions, low detection sensitivity, and low detection rate, and achieve the effects of shortening the detection cycle, high detection sensitivity, and increasing the positive rate
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0040] Example 1 Determination of the specific DNA sequence of Campylobacter coli
[0041]The present invention uses BLASTn and Vector NTI Suite 6.0 software to compare the conservation of DNA sequences of all Campylobacter coli sequencing genes published by NCBI, and screens out the specific sequence of Campylobacter coli genes, the nucleotide sequence of which is as SEQ ID Shown in NO.1.
Embodiment 2
[0042] The design of embodiment 2 campylobacter coli specific primers and probe
[0043] When designing primers and probes in the present invention, attention should be paid to avoiding the mutation point of the sequence. After repeated screening and verification, a pair of fluorescent quantitative PCR primers for detecting Campylobacter coli and the TaqMan® primers used in conjunction with the primers were obtained using Primer Express Version 3 software. A probe, the probe is 5' labeled with a fluorescent group FAM, and 3' labeled with a quencher group BHQ.
[0044] The preferred primer sequence is,
[0045] Upstream primer: CTCGCTTTGGAATCATTCATG, Tm: 57°C,
[0046] Downstream primer: CTTTATTGCCCACAATGATATTTC, Tm: 56°C,
[0047] The preferred probe sequence is,
[0048] FAM-AGGAATCAATGCTGTGGATGAAAATGTAA-BHQ1, Tm: 64°C.
Embodiment 3
[0049] Example 3 Establishment of a fluorescent quantitative PCR detection method for detecting Campylobacter coli
[0050] (1) Optimization of real-time PCR experiment parameters
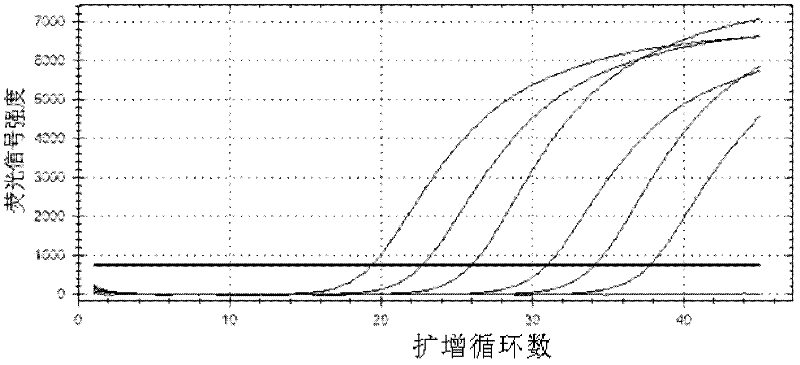
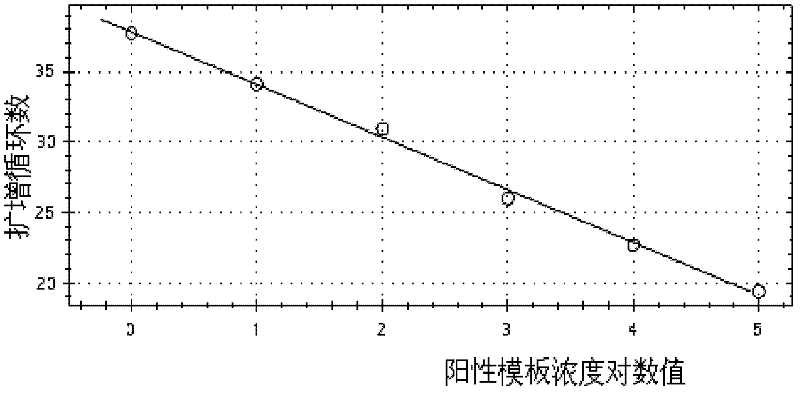
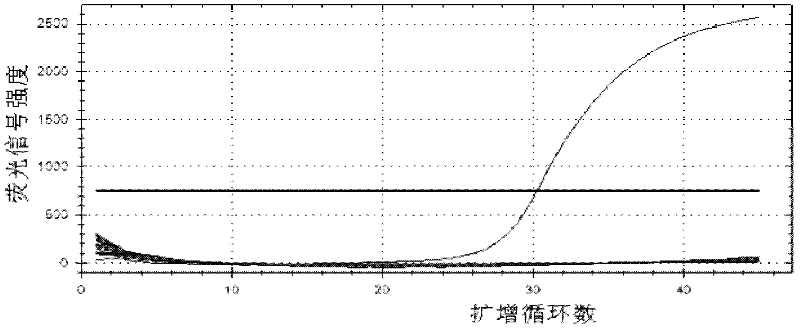
[0051] Refer to Invirogen recommended fluorescent quantitative PCR 25μL system: 2×PCR UDG12.5μL, mg 2+ 4.5mM, PCR nucleotide MIX (including dNTP) 4mM, final concentration of upstream and downstream primers is 0.4μM, final concentration of probe is 0.2μM, DNA template 2μL. Optimize one condition and fix the rest. The optimal conditions were judged according to the Ct value and the strength of the fluorescent signal, and the best conditions were combined as the final optimized system.
[0052] The annealing temperature was optimized from 55°C to 65°C; the primer was optimized according to the final concentration from 0.08μM to 0.62μM; the concentration of the probe was optimized from 0.08μM to 0.36μM; mg 2+ The concentration was optimized between 1.5-5mM; the concentration of dNTPs was optimized b...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com