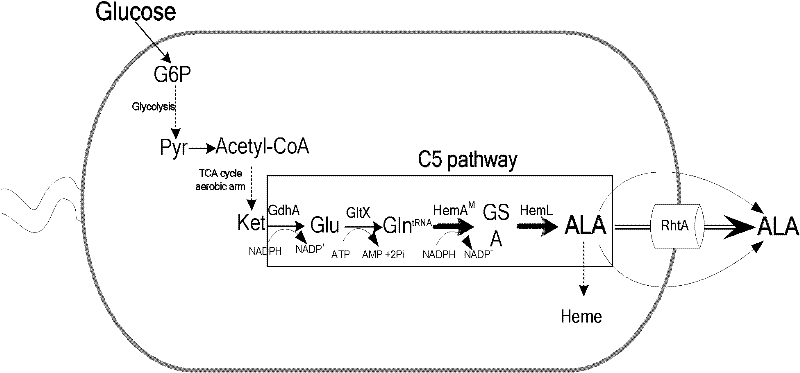
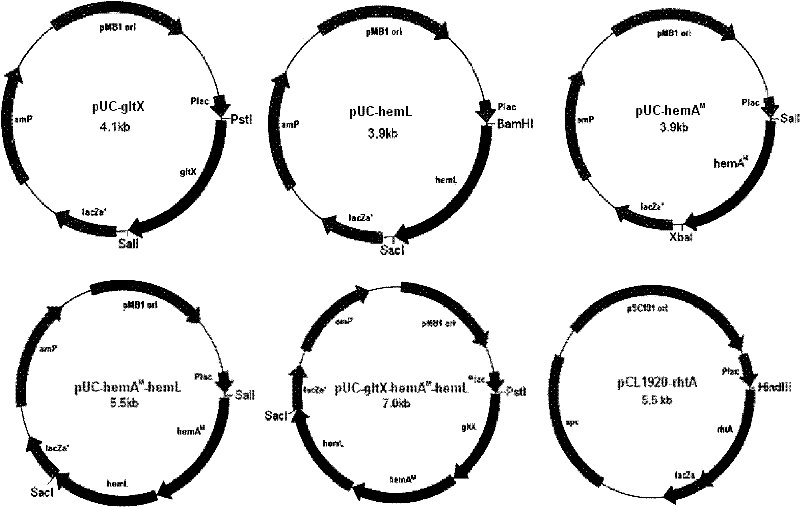
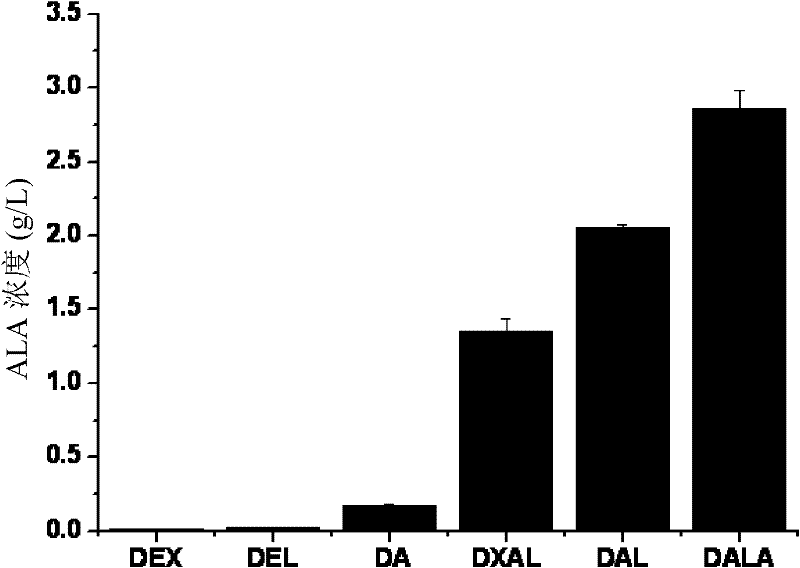
Recombinant escherichia coli and application thereof in production of 5-aminolevulinic acid (ALA)
A technology for recombining Escherichia coli and aminolevulinic acid, which is applied in the fields of genetic engineering and microbial fermentation, can solve problems such as high cost and complex biotransformation process, and achieve important industrial application value
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0050] Embodiment 1, the construction of gltX gene expression vector
[0051] According to the Escherichia coli genome sequence published by NCBI, use the primer gltX-F: 5'-TCC CTGCAG AAAGGAGGATATACATATGAAAATCAAAACTCGCTTCGCGC-3′ and gltX-R:5′-GGC GTC GAC TTACTGCTGATTTTCGCGTTCAGCAATAAAATCC-3' The gltX gene was cloned from the E. coli genome or directly using colony PCR. The cloned gltX fragment was digested with endonucleases PstI and SalI respectively, and the plasmid vector pUC19 was also digested with endonucleases PstI and SalI respectively. The digested gltX fragment and the pUC19 plasmid vector were recovered using an agarose gel kit, and then ligated using T4 ligase. The connection system is 10 μL:
[0052] gltX fragment: 6 μL
[0053] pUC19 vector: 2 μL
[0054] 10×Buffer: 1μL
[0055] T4 ligase: 1 μL
[0056] After ligation at 16°C for 12 h, 10 μL of the ligation solution was transformed into Escherichia coli DH5α competent cells. The transformation process i...
Embodiment 2
[0057] Embodiment 2, the construction of hemL gene expression vector
[0058] According to the Escherichia coli genome sequence published by NCBI, use primer hemL-F: 5′-ACA GGATCC AAAGGAGGATATACATATGAGTAAGTCTGAAAATCTTTTACAGCG-3′ and hemL-R:5′-AAT GAGCTC TCACAACTTCGCAAACACCCGACGTGCAGCA-3' cloned the hemL gene from the E. coli genome or directly by colony PCR. The cloned hemL fragment was digested with endonucleases BamI and SacI respectively, and the plasmid vector pUC19 was also digested with endonucleases BamI and SacI respectively. The digested hemL fragment and the pUC19 plasmid vector were recovered using an agarose gel kit, and then ligated using T4 ligase. The connection system is 10 μL:
[0059] hemL fragment: 6 μL
[0060] pUC19 vector: 2 μL
[0061] 10×Buffer: 1μL
[0062] T4 ligase: 1 μL
[0063] After ligation at 16°C for 12 h, 10 μL of the ligation solution was transformed into Escherichia coli DH5α competent cells. The transformation process is as follow...
Embodiment 3
[0064] Embodiment 3, the mutation of hemA gene and the construction of expression vector
[0065] According to the Salmonella genome sequence published by NCBI, using primer hemA M -F: 5′-CCC GTC GAC AAAGGAGGATATACATATGACCAAGAAGCTTTTAGCACTCGGTATCAAC-3′ and hemA M -R: 5′-AAA TCTAGA CTACTCCAGCCCCGAGGCTGTCGCGCAGA-3′ clone hemA from E. coli genome or directly by colony PCR M Gene. The cloned hemL fragment was digested with endonucleases SalI and XbaI respectively, and the plasmid vector pUC19 was also digested with endonucleases SalI and XbaI respectively. digested hemA M The fragment and pUC19 plasmid vector were recovered using an agarose gel kit, and then ligated using T4 ligase. The connection system is 10 μL:
[0066] hemA M Fragments: 6 μL
[0067] pUC19 vector: 2 μL
[0068]10×Buffer: 1μL
[0069] T4 ligase: 1 μL
[0070] After ligation at 16°C for 12 h, 10 μL of the ligation solution was transformed into Escherichia coli DH5α competent cells. The transformat...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com