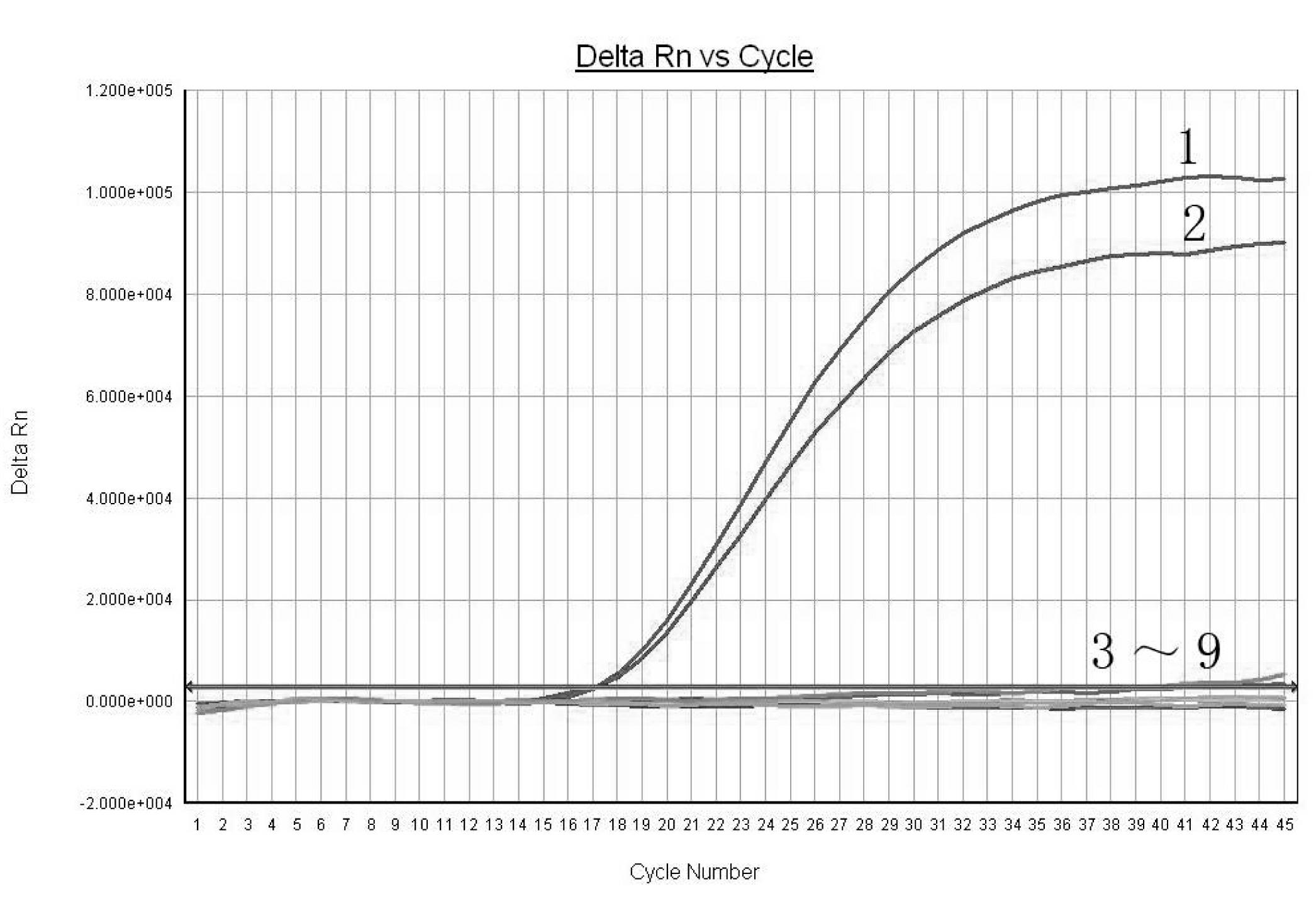
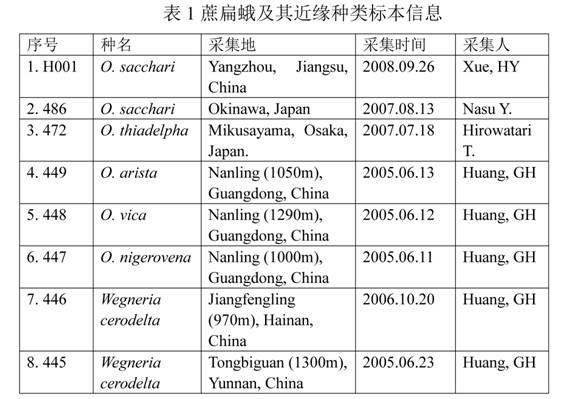
Real-time fluorescence PCR (polymerase chain reaction) detection method based on ITS (internal transcribed spacer) gene for Opogona sacchari
A technology of real-time fluorescence and detection methods, applied in fluorescence/phosphorescence, microbial determination/inspection, biochemical equipment and methods, etc., can solve problems such as time-consuming, difficult operation, and difficulty in obtaining COI genes, and achieve guaranteed Public health safety, accurate and stable detection, and the effect of maintaining ecological balance
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0014] The gene (rDNA) encoding ribosomal RNA is the most conserved structural gene in eukaryotes. It is arranged in the form of concatenated repeats, and each repeat includes three coding regions (respectively encoding 18S, 5.8S and 28S RNA genes), encoding The regions are separated by two ITS (Internal Transcribed Spacers), and the repeats are separated by an ETS (External Transcribed Spacer) and an IGS (Integenic Spacer). Since rDNA has hundreds of copies in the genome (increased Sensitivity of detection) and the intra-species stability and inter-species differences of ITS and IGS regions make it an important object for the study of genetic variation in many organisms, and it is also a hot spot in the research of genetic variation, phylogenetic evolution and molecular identification of insects in recent years , existing studies have shown that the ITS region of many insects in the same group of different species is highly conserved in the length of the base sequence, and the...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com