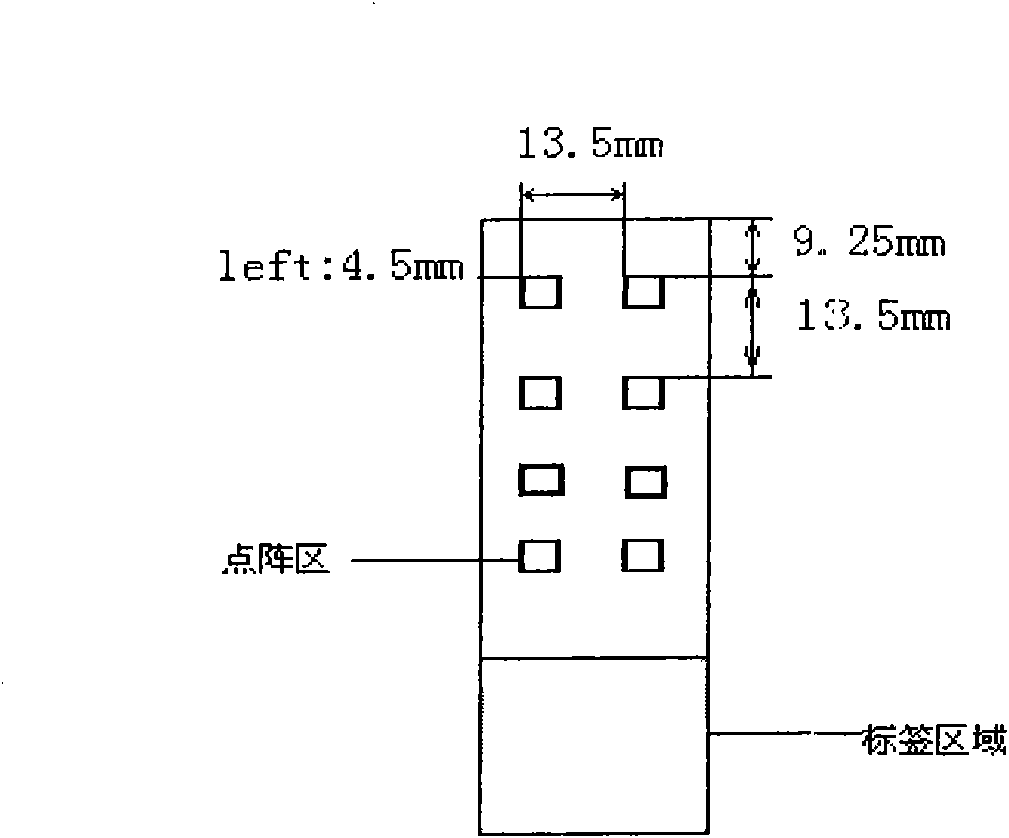
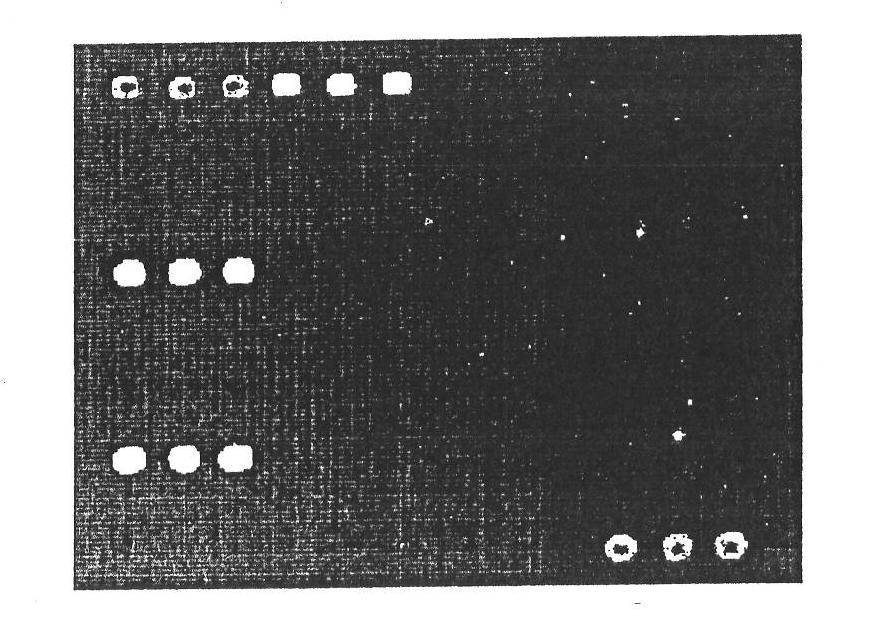
Detection genetic chip and detection kit for infectious diarrhea
An infectious diarrhea, gene chip technology, applied in the field of gene chip application, can solve the problems of inapplicability and increase the workload of detection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0107] Example 1 Probe design and preparation
[0108] 1. Sequence acquisition:
[0109] (1) Bacteria gyrB: Escherichia coli (E.coli) / Shigella (Shigella), Aeromonas hydrophila (Aeromonas hydrophila), Yersinia enterocolitica (Yersinia enterocolitica) were downloaded from the GenBank public database ) of the entire gyrB gene sequence and the entire gyrB gene sequence of its close relatives.
[0110] (2) Acquisition of ITS gene sequences: Download all ITS gene sequences of Salmonella, Vibrio cholerae, Vibrio parahaemolyticus and all ITS genes of their relatives from the GenBank public database sequence.
[0111] (3) Acquisition of dnaJ gene sequences: download all dnaJ gene sequences of Vibrio fluvialis, Vibrio mimicus, Vibrio furnissii and their relatives from the GenBank public database Full dnaJ gene sequence.
[0112] (4) Acquisition of the toxR gene sequence: all toxR gene sequences of Vibriohollisae, Vibrio damsela and their relatives were downloaded from the GenBank ...
Embodiment 2
[0127] Example 2 Primer design and preparation
[0128] 1. Example of primer design:
[0129] (1) Escherichia coli (E.coli) / Shigella (Shigella), hydrophilic Aeromonas (Aeromonashydrophila), enterocolitica (Yersinia enterocolitica) gyrB gene universal amplification primer: the large intestine All the gyrB gene sequences of E.coli / Shigella, Aeromonas hydrophila, and Yersinia enterocolitica were imported into the Glustal X software, and selected A representative sequence was imported into the Primer Primer 5.0 software, and the length was set to 70bp-10bp, the G+C% value was 40%-60%, Hairpin: NONE, Dimer: NONE, False Priming: NONE, Cross Dimer: NONE. And seek out the nucleotide sequence region that is suitable for universal primer design, its characteristic basically meets the following conditions: 1, this constant region should include Escherichia coli (E.coli) / Shigella (Shigella), Aeromonas hydrophilia (Aeromonas hydrophila), gyrB of Yersinia enterocolitica (Yersinia entero...
Embodiment 3
[0146] Example 3 Specific identification of the gene chip
[0147] 1. DNA extraction of target bacteria:
[0148] The collected target bacteria detected by the gene chip and their related strains were cultured overnight for 24 hours; the genomic DNA was extracted with the lysate.
[0149] 2. Amplify the target sequence:
[0150] Take 3 ul of the supernatant extracted by the above genome extraction method as a template and add it to the PCR reaction mixture. The formula of the PCR reaction mixture is shown in Tables 3 and 4 below. (Note: PCR buffer, MgCl2, dNTP mixture in the following tables 3, 4-table 5, 6, Taq enzyme are all purchased from Takara company)
[0151] Table 3 PCR reaction mixture AI formula
[0152]
[0153] Note: ① The concentration of each primer in primer mixture I is: P1, P2, P5, P6 is 0.4 μmol L-1; P13, P14 is 0.2 μmol L-1.
[0154] Table 4 PCR reaction mixture BI formula
[0155]
[0156] Note: ①The concentration of each primer in primer mixture...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com