Molecular reaction system for revealing genetic diversity of tephritidae populations
A technology of genetic diversity and molecular response, applied in the field of molecular response system for revealing the genetic diversity of fruit fly populations, can solve the problems of long quarantine period, unfavorable detection and control of quarantine pests, etc., and achieve good application prospects Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0026] Wherein the preparation of template DNA is as follows:
[0027] Take 5 fruit fly adults from each population and add 800μl extraction buffer (0.1mol / L Tris-HCl, 0.1mol / L NaCl, 0.2mol / L sucrose, 0.05mol / L EDTA, 0.5% SDS), in an ice bath After fully grinding, transfer to 1.5 ml Eppendorf tube, add 0.5mg / ml proteinase K, mix well, put in water bath at 65°C for 1h, add 100μl of 8mol / L KAc, make it fully mix, then ice bath for 1h, 12000r / min at 4°C Centrifuge for 20min, take the supernatant and add 800μl pre-cooled absolute ethanol, mix well, put in ice bath for 2h, centrifuge at 12000r / min for 20min, take the precipitate and wash it once with 70% ethanol, centrifuge at 5000r / min for 5min, and precipitate at 37℃ Dry for 4-6 minutes, finally add 10 μl RNase and 40 μl TE to dissolve, put in water bath at 37°C for 1 hour, and store at 4°C for later use.
Embodiment 1
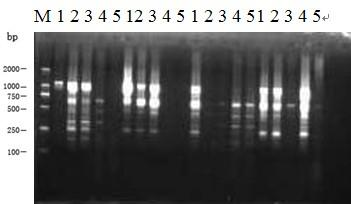
[0029] The fruit flies used in the experiment were mainly collected from the population trapped in the vegetable base in Zhangzhou, Fujian Province. Reagents Buffer (10×PCR Buffer), dNTP, DNAMarker DL 2000, Taq DNA polymerase, etc. required for the experiment were purchased from Shanghai Bioengineering Co., Ltd.; random primers were synthesized by Shanghai Bioengineering Technology Service Co., Ltd.; Tris, HCl, EDTA, NaCl, Kac, ethanol, etc. are domestic analytical reagents. The specific content is as follows:
[0030] 1. Template DNA preparation and validity verification
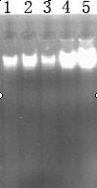
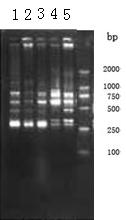
[0031] The template DNA is extracted by using the improved DNA extraction method of the present invention, and then the concentration and purity of the fruit fly DNA are measured by an ultraviolet spectrophotometer and gel electrophoresis to verify the validity of the template DNA.
[0032] (1) Ultraviolet spectrophotometer detection
[0033] Use WFZ800-D3B UV-Vis spectrophotometer to detect the concentr...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com