Fluorescence PCR detection method of pebrine disease and kit thereof
A technology of silkworm microparticles and detection methods, applied in the field of biology
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0043] Example 1: Specimen Collection
[0044] Applicable specimen types include samples such as silkworms and silkworm eggs. Silkworms, take the suspected diseased silkworms and transfer them to a 5ml centrifuge tube, seal them and submit them for inspection. For silkworm eggs or excrement, about 1 g of silkworm eggs or excrement from suspected diseased silkworms should be taken, transferred to a 1.5ml centrifuge tube, sealed and sent for inspection.
Embodiment 2
[0045] Embodiment 2: the extraction of silk microsporidia DNA
[0046] Take the above specimen and add an appropriate amount of normal saline to grind, filter the homogenate with gauze, collect the filtrate, and use differential centrifugation (500rpm, centrifuge for 2 minutes, discard the precipitate, and then centrifuge the supernatant at 5000rpm for 10 minutes) to discard the supernatant. Suspend with an appropriate amount of physiological saline to obtain a crude extract of Nb spores. Take 0.5ml (10 8 individual / ml) Microsporidia suspension, 5000rmp, centrifuge for 5 minutes, remove the supernatant, keep the spores, add 0.2mol / LKOH 0.25ml at the same time, mix well, induce at 27℃ for 1 hour, centrifuge at 5000rpm for 5 minutes, remove the supernatant . Resuspend with 0.5ml of normal saline and place at 25°C for 30 minutes. Centrifuge at 5000rpm for 5 minutes, add 50μl of nucleic acid extraction solution to the precipitate and mix thoroughly, then bathe in boiling water ...
Embodiment 3
[0047] Embodiment 3: the amplification detection of silk microsporidia DNA
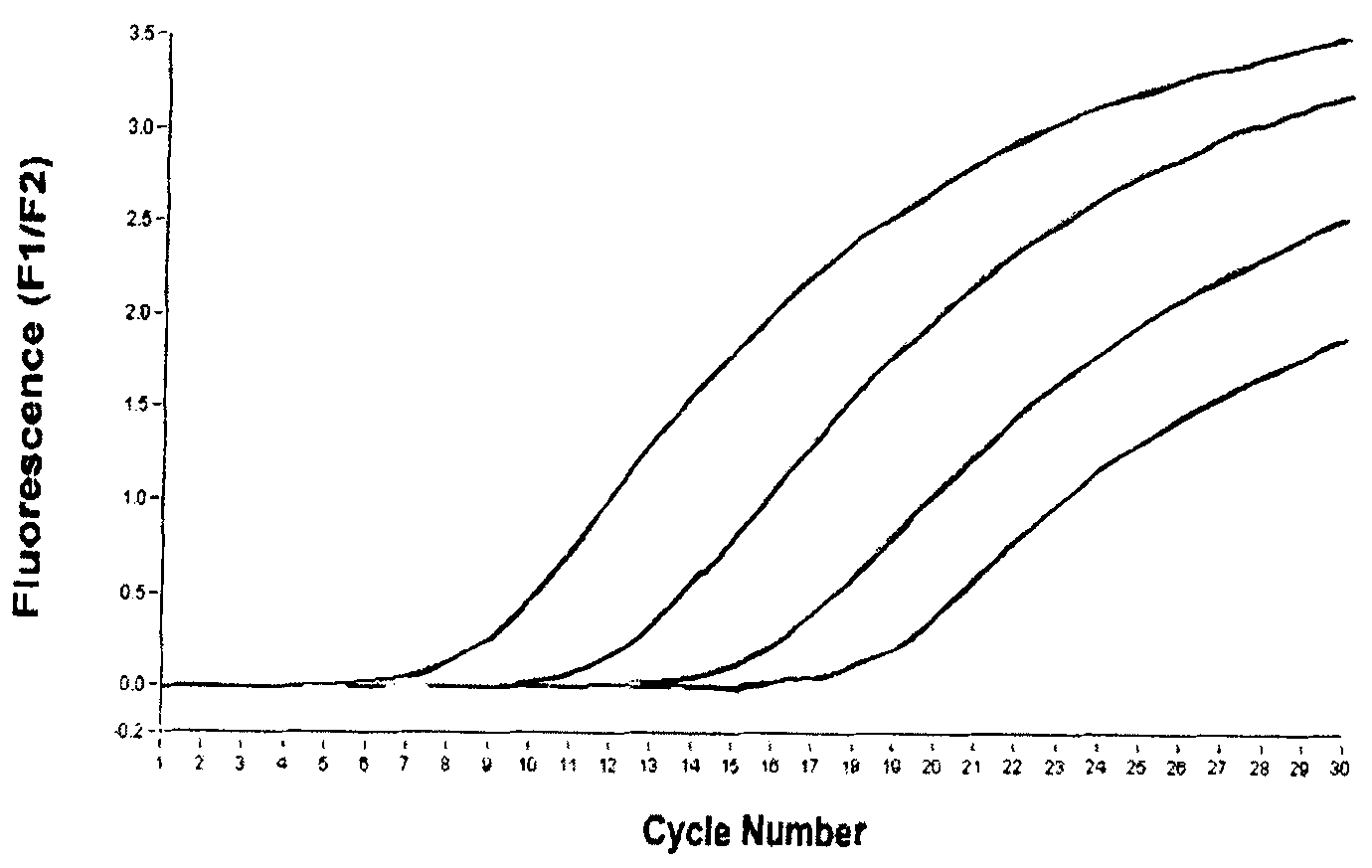
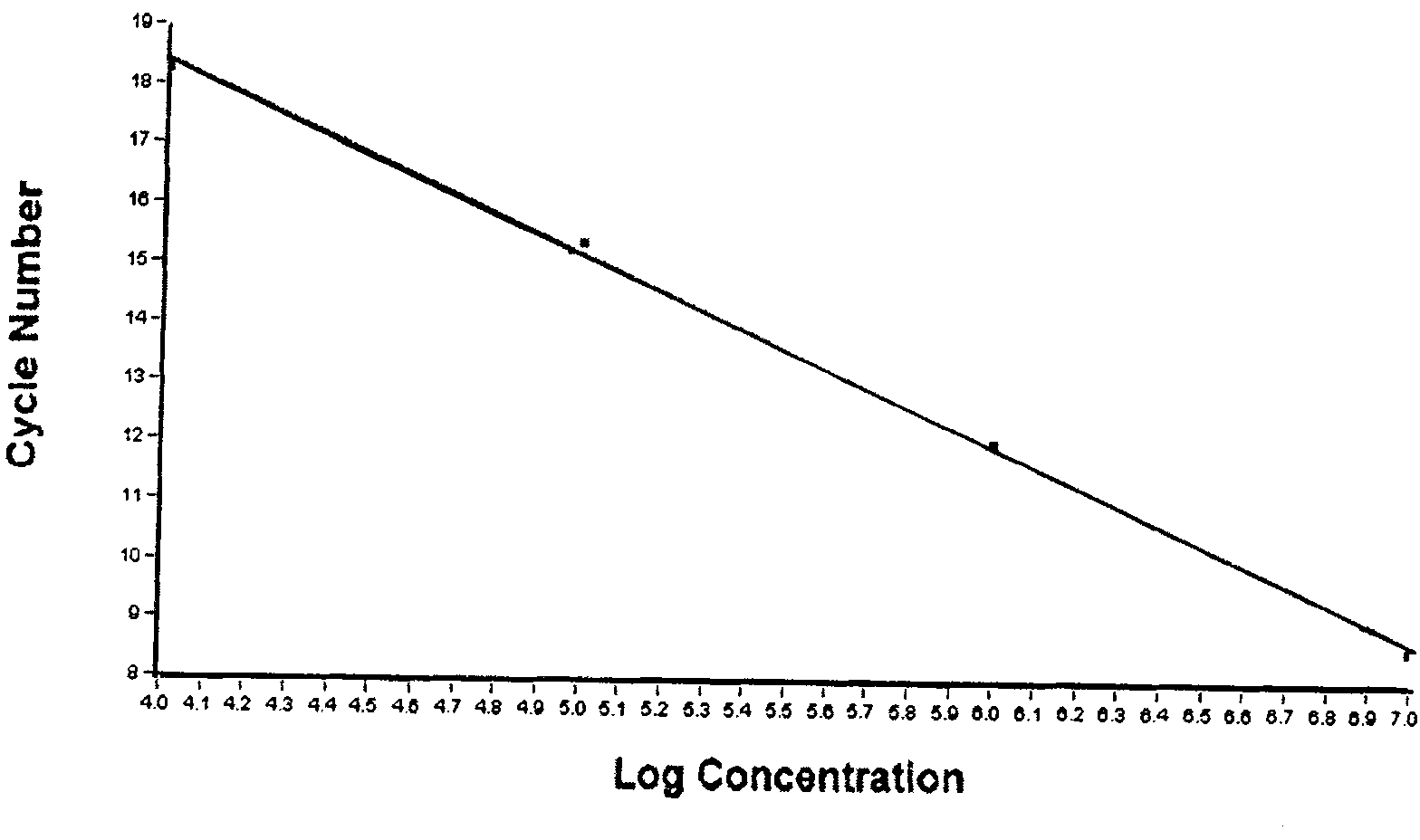
[0048] Take out the NB-PCR MIX and Taq enzyme system from the kit and wait for room temperature to melt, shake and mix well, then centrifuge at 10,000rpm for 10 seconds. Each test reaction system was prepared as follows, PCR MIX 24μl, Taq enzyme system 2ul, put into a clean 0.2ml PCR tube, mix well, and centrifuge at 10,000rpm for 10 seconds. Add 4 μl of processed sample (extracted DNA) or positive quality control or negative quality control to the set PCR reaction tube, and centrifuge briefly at 10,000 rpm for 10 seconds.
[0049] Put each reaction tube into the reaction tank of the quantitative PCR instrument, set the name of each detection, the type of fluorescent group (FAM is selected as the reporter group, TAMRA is selected as the quencher group) and amplification conditions: ABI PRISM 7700, ABI PRISM5700, The amplification conditions of ABI GeneAmp 7000, ABI PRISM7300 / 7500, MJ Opticon and othe...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com