Kit and method for detecting Microsporidium silkworm by pcr-elisa method
A technology of microsporidia and kits, applied in the field of detection, can solve the problems of high cost, high requirements, confusion, etc., and achieve the effect of high specificity, specificity and accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
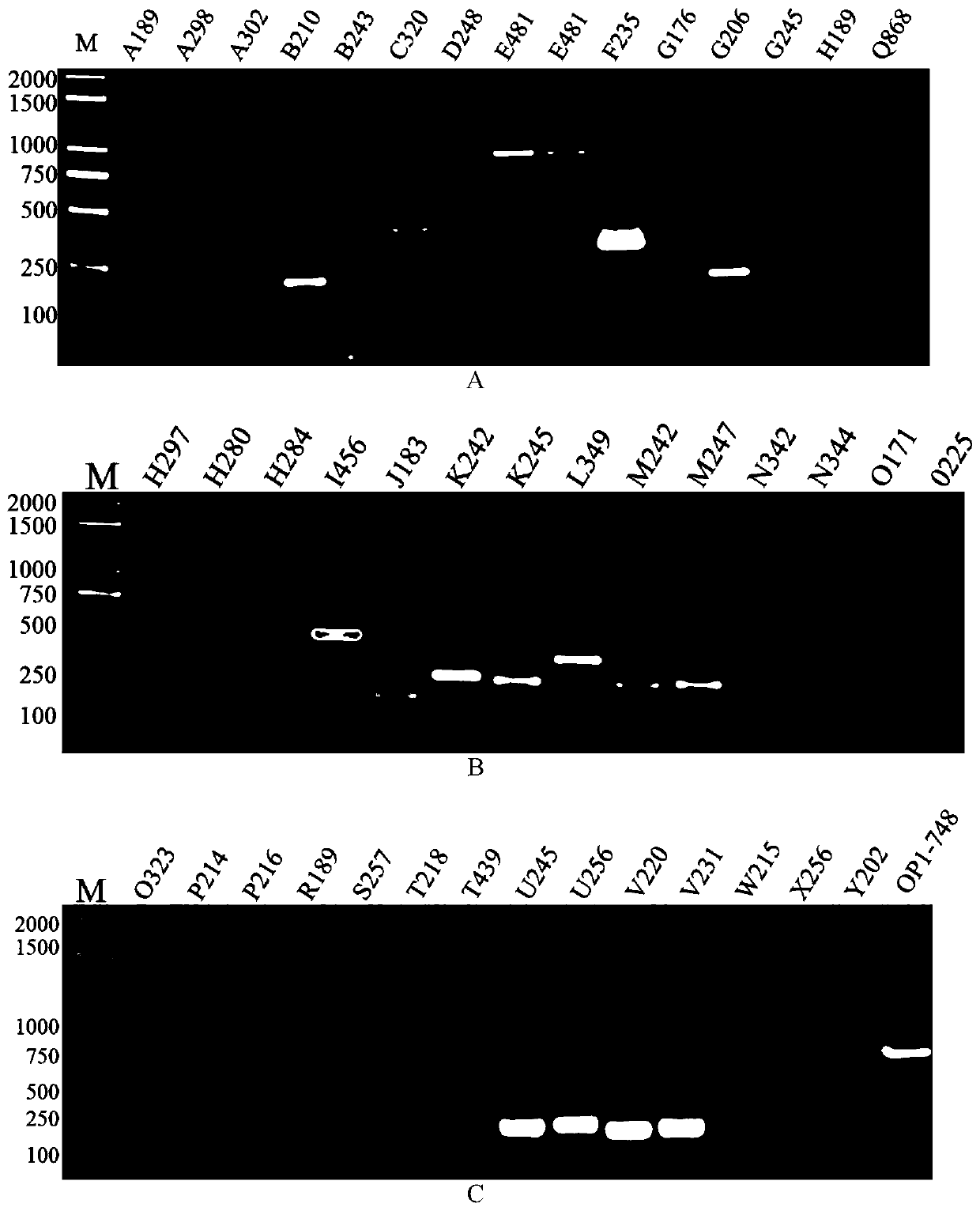
[0047] Embodiment 1, primer screening
[0048] Genomic DNA of N. silkworm was extracted by CTAB method.
[0049] At the same time, the genomes of Nos. tussah, No. bee and No. naka proteus were extracted as controls.
[0050] Then according to the comparison between the genome of Microsporidium silkworm and several other Microsporidium genomes, the sequences with similarity less than 30% were screened out for primer design.
[0051] The specific primers are shown in Table 1 below.
[0052] Table 1. Primer pairs for microsporidia detection
[0053]
[0054]
[0055]
[0056] With the silkworm Microsporidia genomic DNA as a template, the sequences shown in Table 1 are primers for PCR amplification, and the PCR amplification system is as shown in Table 2 below:
[0057] Table 2. PCR amplification system
[0058]
[0059]
[0060] Amplify according to the following conditions, pre-denaturation at 98°C for 2 min; denaturation at 98°C for 10 sec, annealing at 64°C ...
Embodiment 2
[0061] Embodiment 2, specificity experiment
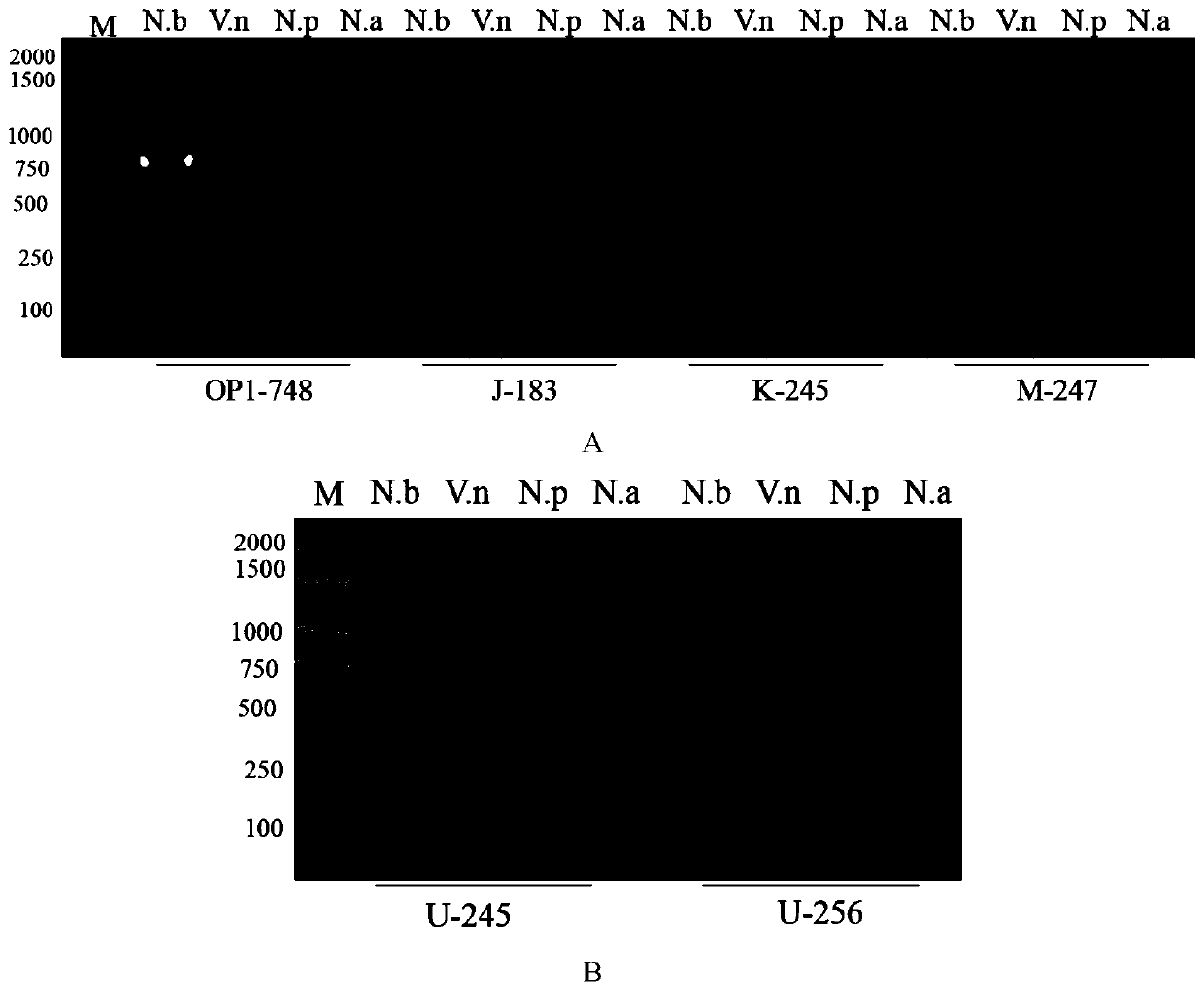
[0062] According to the method of Example 1, the six pairs of primers screened are used to detect the following templates respectively: 1. DNA of Bombyx mori No. spp.; 2. No. naka proteus DNA; The company's MightyAmp DNA polymerase Ver.3, the total reaction system is 25μL. Amplification was performed with an Applied Biosystems PCR instrument, and the reaction parameters were: pre-denaturation at 98°C for 2 minutes; denaturation at 98°C for 10 sec, annealing at 64°C for 15 sec, extension at 68°C for 35 sec, and 30 cycles; final extension at 68°C for 3 min, and storage at 12°C. Take 10 μL of the amplified product for electrophoresis in 2% agarose gel electrophoresis, then stain with ethidium bromide, and observe under ultraviolet light to determine whether there are specific bands. The results are as follows: figure 2 shown in . The results show that OP1-748, K245, M247, U245, and U246 five pairs of primers can specifically amplif...
Embodiment 3
[0063] Embodiment 3, false positive detection
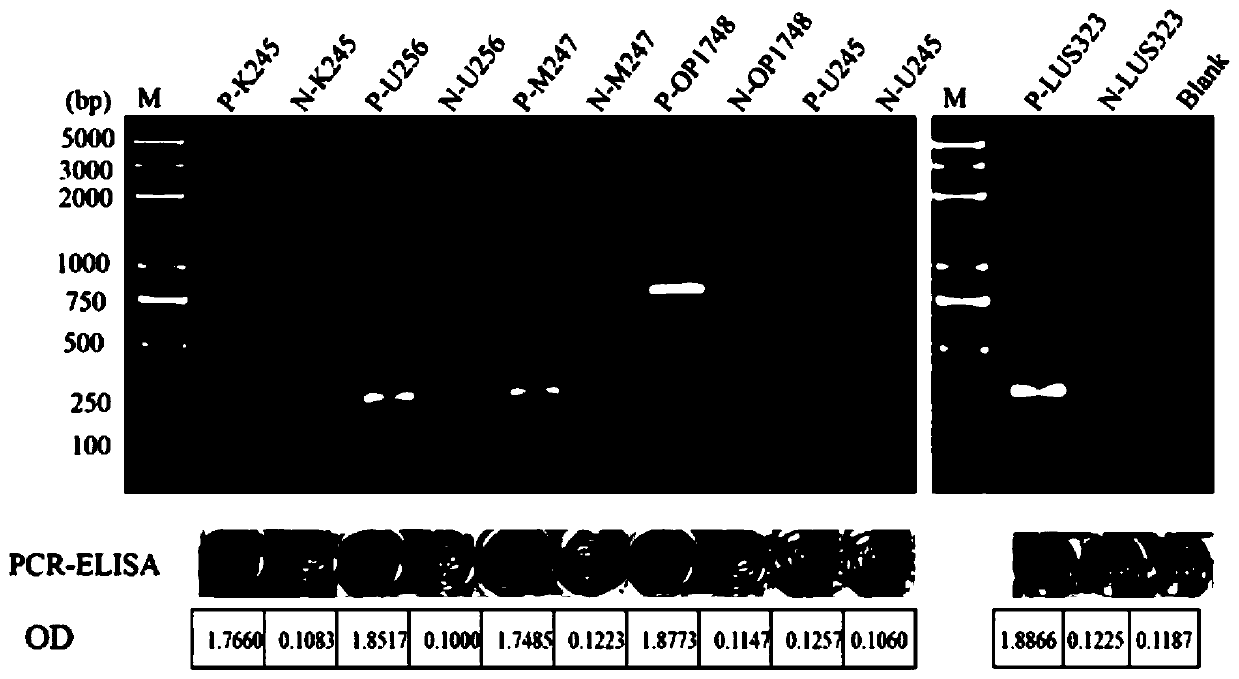
[0064] In order to detect whether there are false positives in the labeled primers, DNA of Bombyx mori was used as a template for positives, and ddH was used for negatives 2 O was used as a template and amplified according to the established PCR system. After amplification, 10 μL was used for agarose gel electrophoresis. The conditions of gel electrophoresis are: 1×TAE buffer, voltage 180V, electrophoresis time 30min. At the same time, it was detected by ELISA, and the results were as follows: image 3 shown. The results showed that except for U245, which could not get positive results, OP1-748, K245, M247, U246, and LSU323 could all get positive results, and the OD of the negative control was close to that of the blank control, so the next step of the experiment could be carried out. For the specific sequence of LSU323, please refer to the literature (Ni Qi, Establishment of a lateral flow test strip detection method for the...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com