Antimicrobial peptide gene from pinctada fucata and application
A technology of Hepu pearl oyster and antibacterial peptide, applied in the field of genetic engineering, can solve problems such as high mortality and achieve the effect of great economic value
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0060] Example 1: Hepu Pinctada cDNA library construction and EST screening
[0061] The total RNA ( Figure 1a ),use Synthesis kit (Stratagene), Gigapack III Gold Cloning kit (Stratagene) and Kits such as mRNA isolation system III (Promega) were used to construct EST libraries ( Figure 1b ). Randomly pick single clones, use primer T 3 (AATTAACCCTCACTAAAGGG) was sequenced, and the carrier was removed through the Crossmatch program; through the Phrap program, the splicing and clustering was performed to obtain the UniGene data set. After the Phrap program was processed, the EST sequences that could be gathered together were Contigs sequences, and the EST sequences that could not be gathered together were Singlets sequence, the two parts of the sequence are combined to obtain the UniGene data set after EST de-redundancy. The obtained Contigs and Singletons were analyzed by BLASTn and BLASTx in the database, and one of the EST sequences was identified to have high homol...
Embodiment 2
[0062] Example 2: PCR amplification of Hepu Pinctada cDNA library
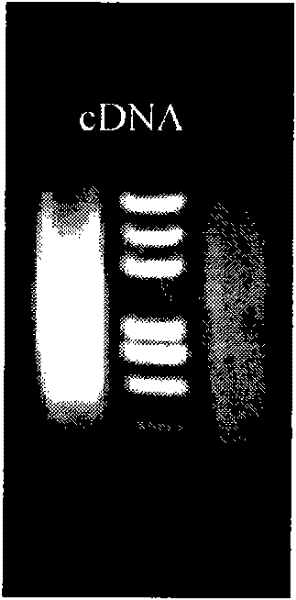
[0063] Using the Hepu Pinctada cDNA obtained by reverse transcription with the GeneRacer Kit as a template, the primers were the 5'-Primer and 3'-Primer of the GeneRacer Kit, respectively, and amplified by touchdown PCR to establish the Hepu Pinctada cDNA library. The reaction conditions were as follows: Pre-denaturation at 94°C for 5 minutes, followed by 40 cycles, divided into two stages: ① Denaturation at 94°C for 30 seconds, decrease by 0.5°C per cycle from 68°C to 54°C, annealing for 30 seconds, extension at 72°C for 2 minutes, a total of 32 cycles; ② 94°C Denaturation for 30S, annealing at 50°C for 30S, extension at 72°C for 2min, a total of 8 cycles. Finally, extend at 72°C for 10 min. Electrophoresis of PCR amplification products figure 2 .
Embodiment 3
[0064] Example 3: PCR amplification of the 3' end of the antimicrobial peptide from Hepu Pearl Oyster
[0065] The operation was carried out according to the instructions of GeneRacer Kit. According to the requirements of the kit, two upstream primers GSP1 and GSP2 were used for nested PCR according to the sequenced EST sequence of the antimicrobial peptide of Hepu pinctadae. The first upstream primer (GSP1) is 20 bases (5'-TGTGAATGCCTGGTGGTCTA-3') from the 96th base of the sequenced Hepu pearl oyster antimicrobial peptide EST sequence, and the second upstream primer (GSP2) is 20 bases from position 164 (5'-GCCGTGCTGCTACTGGTTAT-3').
[0066] For the first PCR, the PCR product dilution product (1:100) obtained in the above-mentioned embodiment 2 was used as a template, and the primer was GSP1, and the nucleotide sequence from the 96th to the 3'-polyA end was amplified by PCR, and the PCR method of Eppendorf was used. The PCR amplification program was set on the instrument, an...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com