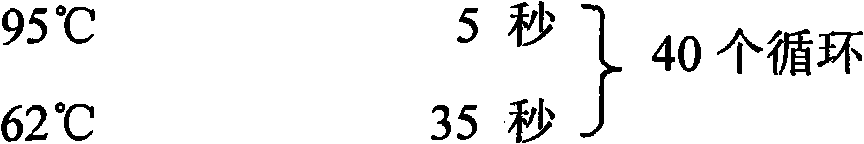Trypsin gene cDNA sequence of Scylla paramamosain and cloning method and application thereof
A technology of trypsin gene and mud crab, which is applied in the field of cDNA sequence and cloning of the trypsin gene of mud crab, can solve the problem of estimating the amount of feeding, optimize the seedling breeding process, improve the survival rate of seedlings, The effect of reducing bait consumption
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
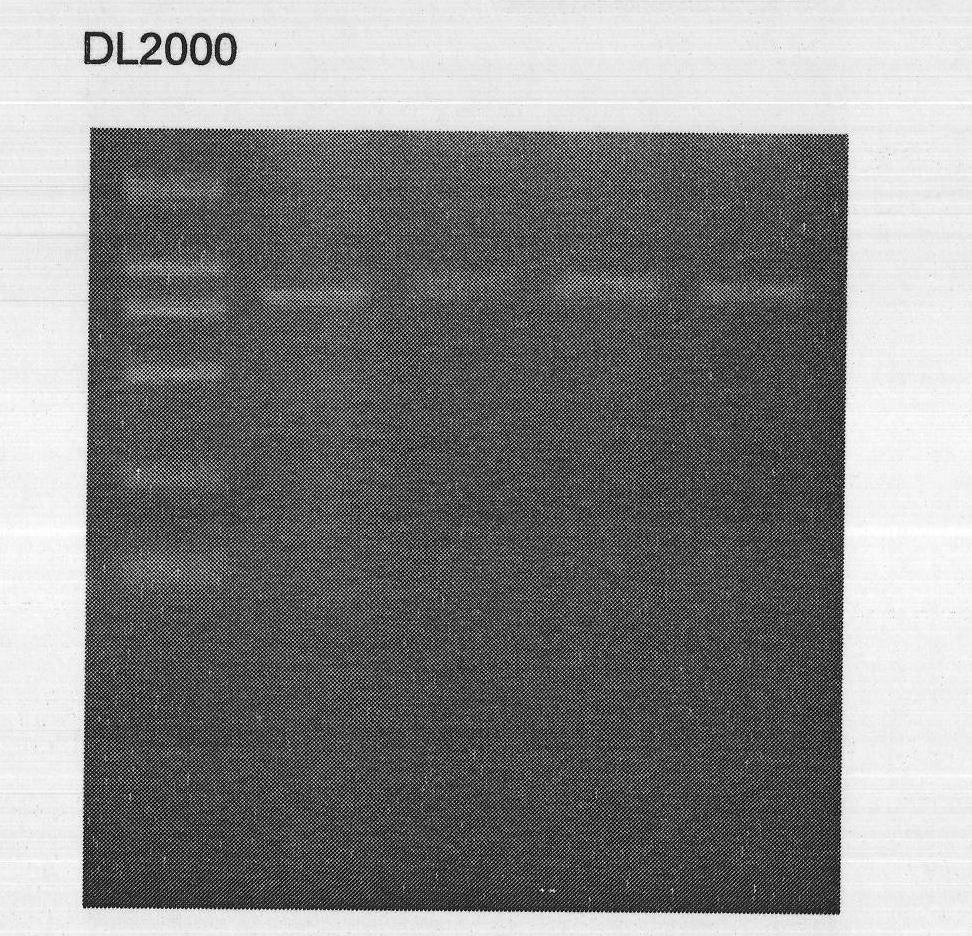
[0036] Cloning of trypsin gene from Scylla syringae
[0037] (1) RNA isolation (RNA isolation)
[0038] Take the hepatopancreas of Scylla pseudocavena, add liquid nitrogen, grind it with a mortar, add it to a 1.5mL EP tube filled with Trizol lysate, oscillate fully, extract total RNA (TRIzol Reagents, TaKaRa), and use formaldehyde deformable gel electrophoresis Identify the quality of total RNA, and then measure the RNA content on a spectrophotometer;
[0039] Proceed as follows:
[0040] 1. Homogenization
[0041] Add 10-30mg of tissue to 1ml TRIzol, and use an electric homogenizer or disposable grinding pestle to fully homogenize;
[0042] 2. Phase Separation
[0043] a. After homogenization, place at room temperature for 5 minutes to fully lyse the sample, then centrifuge at 12,000 rpm for 10 minutes at 4°C, and take the supernatant;
[0044] b. Then add 200 μl of chloroform, vibrate vigorously and mix well, then place at room temperature for 3-5min to allow natural ph...
Embodiment 2
[0069] Fluorescence Quantitative PCR Detection of Trypsin Gene in Scylla sclerophyllum
[0070] Application in Predicting Feed Amount and Moulting Cycle of Scylla Seedlings
[0071] (1) Total RNA extraction
[0072] Take individual blue crabs at each daphnia stage, add liquid nitrogen, grind with a mortar, add 1.5mL EP tube containing lysate, shake fully, extract total RNA (TRIzol Reagents, TaKaRa), use formaldehyde deformable gel Identify the quality of total RNA by electrophoresis, and then measure the RNA content on a spectrophotometer;
[0073] (2) Fluorescent quantitative PCR experiment
[0074] According to TaKaRa's SYBR According to the instructions of ExScript TM RT-PCR Kit (Perfect Real Time), the reverse transcription conditions are 42°C for 15 minutes and 95°C for 2 minutes;
[0075] Take the reverse-transcribed cDNA extracted from samples of each daphne larvae stage of Scylla syringae, and take 2 μl and dilute it with EASY Dilution (TaKaRa Code: D9160, Shiga, ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com