Primer pairs for detecting germs of paddy bacterial glume blight and detection method
A kind of rice blight bacteria, bacterial technology, applied in the field of primers and detection for the detection of rice bacterial rice blight, can solve the problems of lack of detection methods and standards for rice bacterial blight, achieve high accuracy, high detection sensitivity, Good effect of primer specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0026] Primer design and synthesis
[0027] According to the reported ITS region sequence (genebank D87080) of bacterial rice blight of rice, the primer sequence was designed as follows:
[0028] SEQ ID NO.1 (forward): 5'-ACACGGAACACCTGGGTA-3';
[0029] SEQ ID NO. 2 (reverse): 5'-TCGCTCTCCCGAAGAGAT-3'.
[0030] According to the reported gryB gene (genebank AB207074) of bacterial rice blight of rice, primers and probes were designed, and the primer sequences are:
[0031] SEQ ID NO.3 (forward): 5'-GCAGCGGCAAGGAAGACG-3';
[0032] SEQ ID NO. 4 (reverse): 5'-GTCGTCGCCCGACGTCTC-3'.
Embodiment 2
[0034] Isolation of Rice Bacterial Rice Blight from Hybrid Rice Seeds
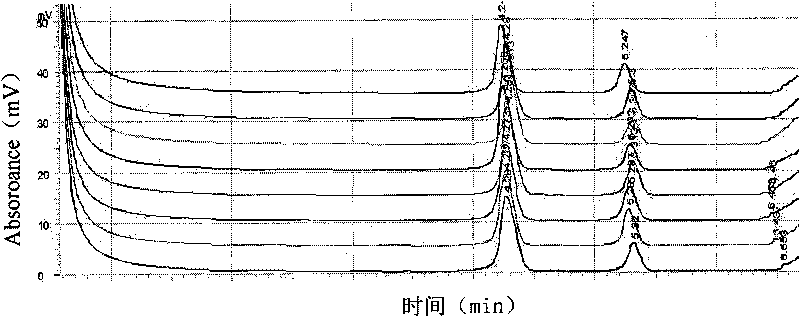
[0035]The semi-selective isolation medium S-PG agar medium reported in the literature was used to isolate and cultivate the pathogen in rice materials. The bacterial grain blight showed two growth forms on S-PG, one colony was reddish-brown, round, smooth, and raised; the other colony was lavender, round, smooth, and raised.
[0036] Sample handling method
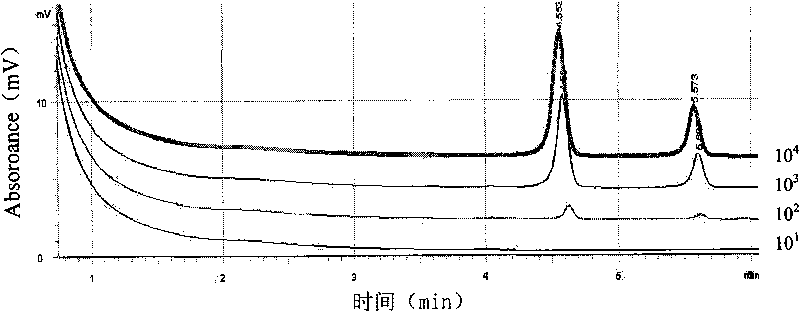
[0037] After washing a large number of seed samples with distilled water, weigh 10g or 400 seeds and put them into 90mL 0.001% Tween-phosphate buffer (pH7.0) for 4-6 hours at low temperature (5°C-15°C), The instrument was smashed to prepare the sample extract. If it is to detect a small amount of seed samples, take 20 seeds and add 10 mL of sterilized 0.001% Tween-phosphate buffer (pH 7.0), fully grind the seeds with a sterilized mallet and mortar or agitator, and prepare into an extract.
[0038] Place the sample extract in an environment of 22°C-...
Embodiment 3
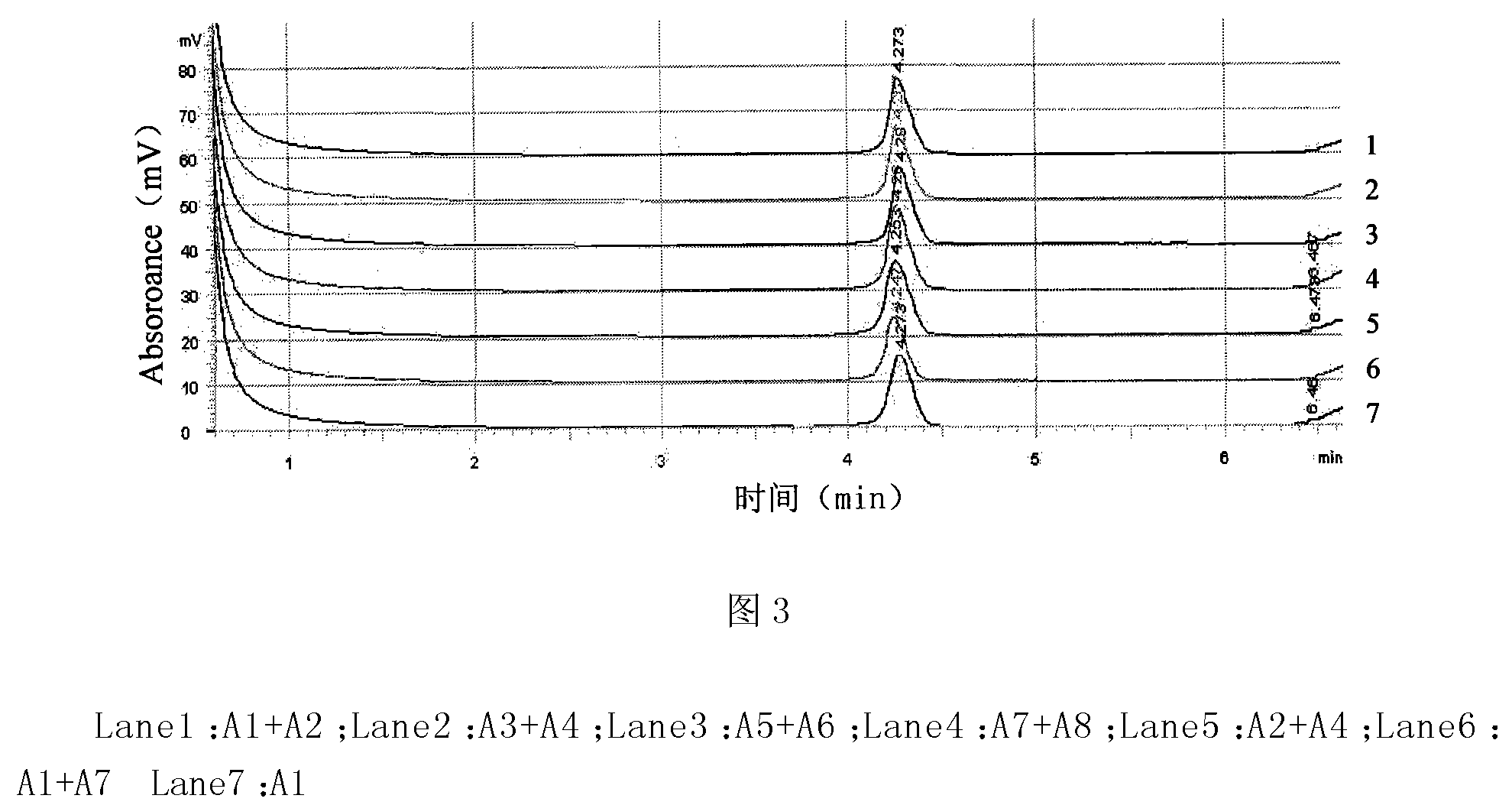
[0041] Bacterial DNA Extraction
[0042] 1) Directly pick at least 5 typical or suspected colonies grown on the S-PG plate into a 1.5mL centrifuge tube, add 1mL sterilized water to shake fully, centrifuge at 12000g for 3min, discard the supernatant, and then add 1mL sterilized Water, repeat three times, take the precipitate for DNA extraction;
[0043] 2) Add 600 μL of TE buffer, 30 μL of 10% SDS solution, and 15 μL of 20 mg / mL proteinase K to the precipitate, mix well, and incubate in a water bath at 37° C. for 1 h.
[0044] 3) Add an equal volume of chloroform-isoamyl alcohol (24:1), and mix well. Centrifuge at 10,000 g for 5 min, and transfer the supernatant to a new centrifuge tube.
[0045] 4) Add an equal volume of phenol-chloroform-isoamyl alcohol (25:24:1), mix well, centrifuge at 10000 g for 5 min, and transfer the supernatant to a new centrifuge tube.
[0046] 5) Add 0.6 times the volume of isopropanol, mix gently, precipitate at -20°C for 1 hour, centrifuge at 10...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com