Method for rapidly, qualitatively and quantitatively detecting microcystic aeruginosa on spot and the reagent kit
A technology for quantitative detection of Microcystis aeruginosa, which is applied in the directions of microorganism-based methods, biochemical equipment and methods, and microorganism determination/inspection, can solve the problems of purifying algal cells and long detection time, and achieves simple and accurate operation. high sex effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0046] Example 1: A kit and method for on-site rapid qualitative and quantitative detection of Microcystis aeruginosa
[0047] The present invention uses the transcription unit spacer ITS (Internal Transcribed Spacer) between the ribosome subunits of Microcystis aeruginosa as the target sequence, and searches for a highly specific DNA sequence in the target fragment as a probe. The key to the qualitative analysis of microalgae in natural water blooms using whole-cell fluorescence in situ hybridization is to find suitable DNA fragments in the genome of Microcystis aeruginosa as probes. This DNA segment must be highly variable, that is, the sequence behaves differently between different species and even between different geographical strains of the same species. ITS sequences can be considered as FISH probes due to their high variability.
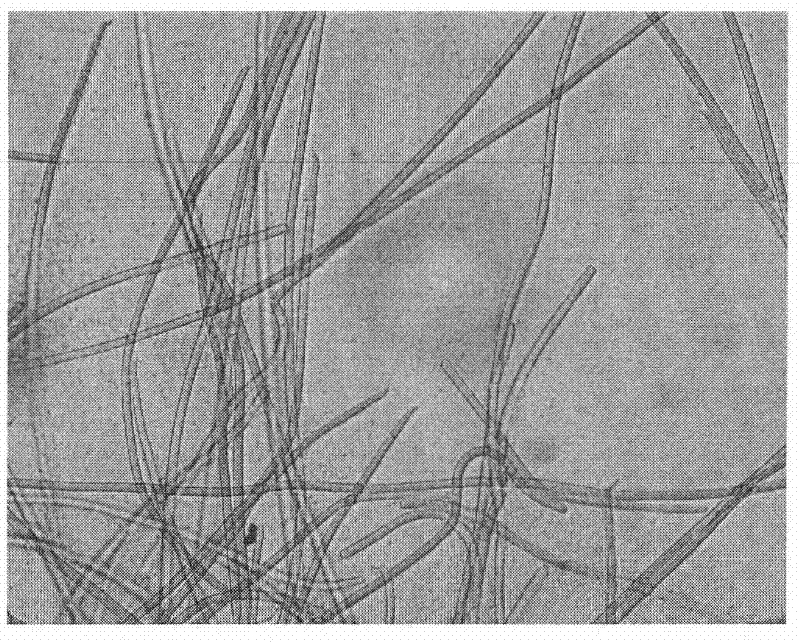
[0048] Microcystis aeruginosa (photographs under natural light conditions are attachedfigure 1 (shown) is the main species of bloom microal...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com