Method for constructing encephalomyocarditis virus infections clone
A technology of encephalomyocarditis virus and infectious cloning, applied in the field of genetic engineering, can solve problems such as limitations in molecular virology research, achieve strong controllability, increase success rate, and ensure fidelity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
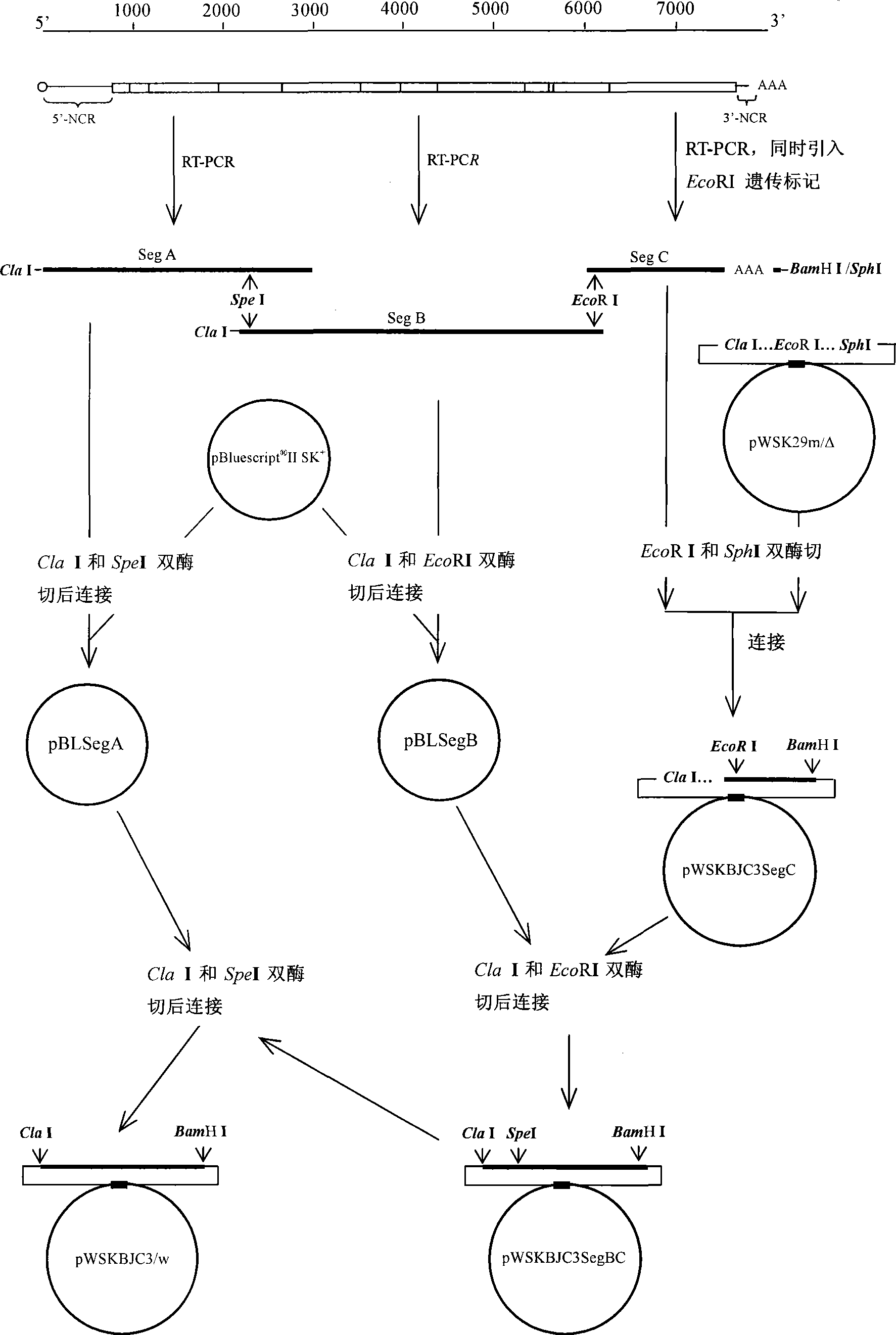
[0024] Embodiment 1 Transformation of low copy plasmid vector
[0025] According to the complete sequence of the low copy plasmid vector pWSK29 and Instructions for MultiSite-directed Mutagenesis Kit (Stratagene, Denmark), design 1 pair of palindromic primers Spe-Upper and Spe-Lower to mutate the Spe I site at position 3536, and design 1 pair of palindromic primers BssH-Upper and BssH-Lower The BssH II site at position 5124 was mutated. Design primers based on a nonsense sequence of the PRRSV Nsp2 gene, add BssH II and Cla I restriction sites to the 5' end of the upstream primer pNSP2-F, and sequentially add the 5' end of the downstream primer pNSP2-R BssH II and Sph I two enzyme cutting sites, with HS DNA Polymerase (Takara, Dalian) amplified this fragment and digested it with BssHII. At the same time, the mutated pWSK29 vector was digested with BssHII, and the digested product was dephosphorylated, and then T4 ligase (Promega, USA ) Ligate the above-mentioned amplifie...
Embodiment 2
[0029] Example 2 Extraction of encephalomyocarditis virus total RNA
[0030] The F8 generation cells of porcine encephalomyocarditis virus BJC3 were used to culture the virus, and the total RNA of the BJC3 virus was extracted according to the specific steps in the kit manual using the QIAampViral RNA Mini Kit (Qiagen, Germany). The obtained RNA was dissolved in 20 μL RNase-free water (Ambion Inc., USA), and frozen at -70°C.
Embodiment 3
[0031] Example 3 RT-PCR method for segmental amplification of the full-length cDNA of encephalomyocarditis virus
[0032] According to the whole genome sequence of porcine encephalomyocarditis virus BJC3 (GenBank No. DQ464062), design 3 pairs of primers covering the whole genome of the virus, the primer sequences and their positions in the BJC3 genome are as shown in Table 2: the upstream primers for full-length amplification T7 RNA polymerase promoter core sequence is added to T7-SegA-F to ensure high abundance and high quality RNA during in vitro transcription; Cla I restriction site is added to T7-SegA-F and ClaI-SegB-F ; Sph I and BamH I two enzyme cutting sites are added to SegC-R; Point mutations are introduced into primers SegB-R and SegC-F, and after amplification, T at position 6262 of the genome is mutated to A, and the codon after mutation Both CGA and the pre-mutation codon CGT encode arginine, which is a synonymous mutation. The obtained sequence GAATTC is the rec...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com