Process for quickly detecting DNA methylation
A methylation and rapid technology, applied in the field of medical biological detection, can solve the problems of long time for dialysis desalination, heavy workload, complicated operation, etc., and achieve the effect of saving desalination time, short detection time and improving repeatability.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
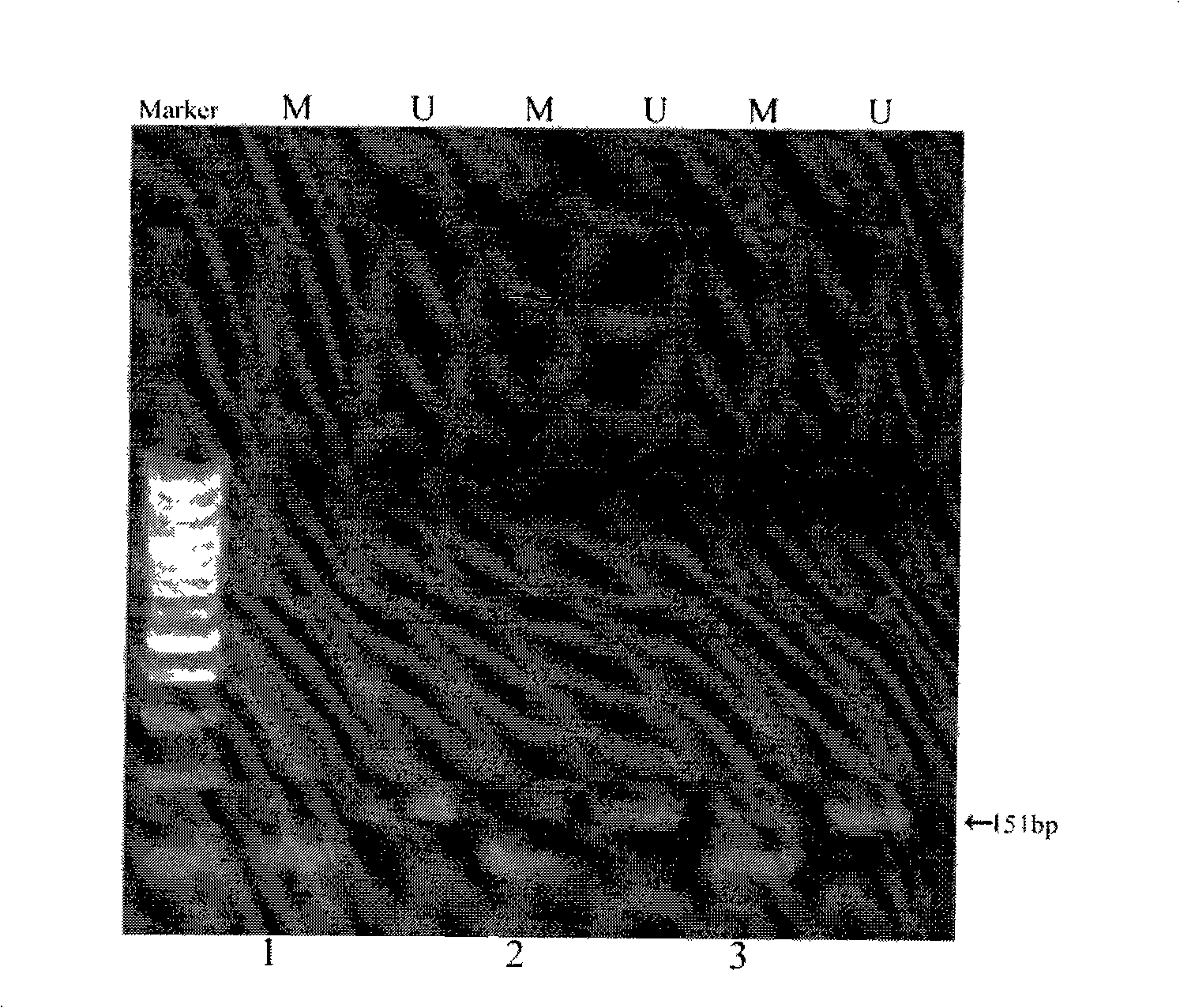
[0071] Example 1 Using the method of the present invention to detect the methylation of the p16 gene in liver cancer specimens
[0072] The samples to be tested were taken from individual samples of 3 liver cancer patients in Shanghai Oriental Hepatobiliary Hospital. The p16 gene has been shown to be hypermethylated in liver cancer (Naoyuki Umetani, MichielF.G.de Maat, Eiji Sunami, Suzanne Hiramatsu, Steve Martinez, and Dave S.B. Hoon Methylation of p16 and Ras Association Domain Family Protein 1a during Colorectal Malignant Transformation), However, liver cancer cannot be diagnosed by the hypermethylation of a gene, and not all patients with liver cancer will have hypermethylation of the p16 gene, and the methylation of the p16 gene is only one of the reference indicators. The operation steps are as follows (the three samples have the same method):
[0073] 1. Prepare the DNA to be tested:
[0074] Take 50 mg of fresh liver tissue, and extract DNA with a genomic DNA extract...
Embodiment 2
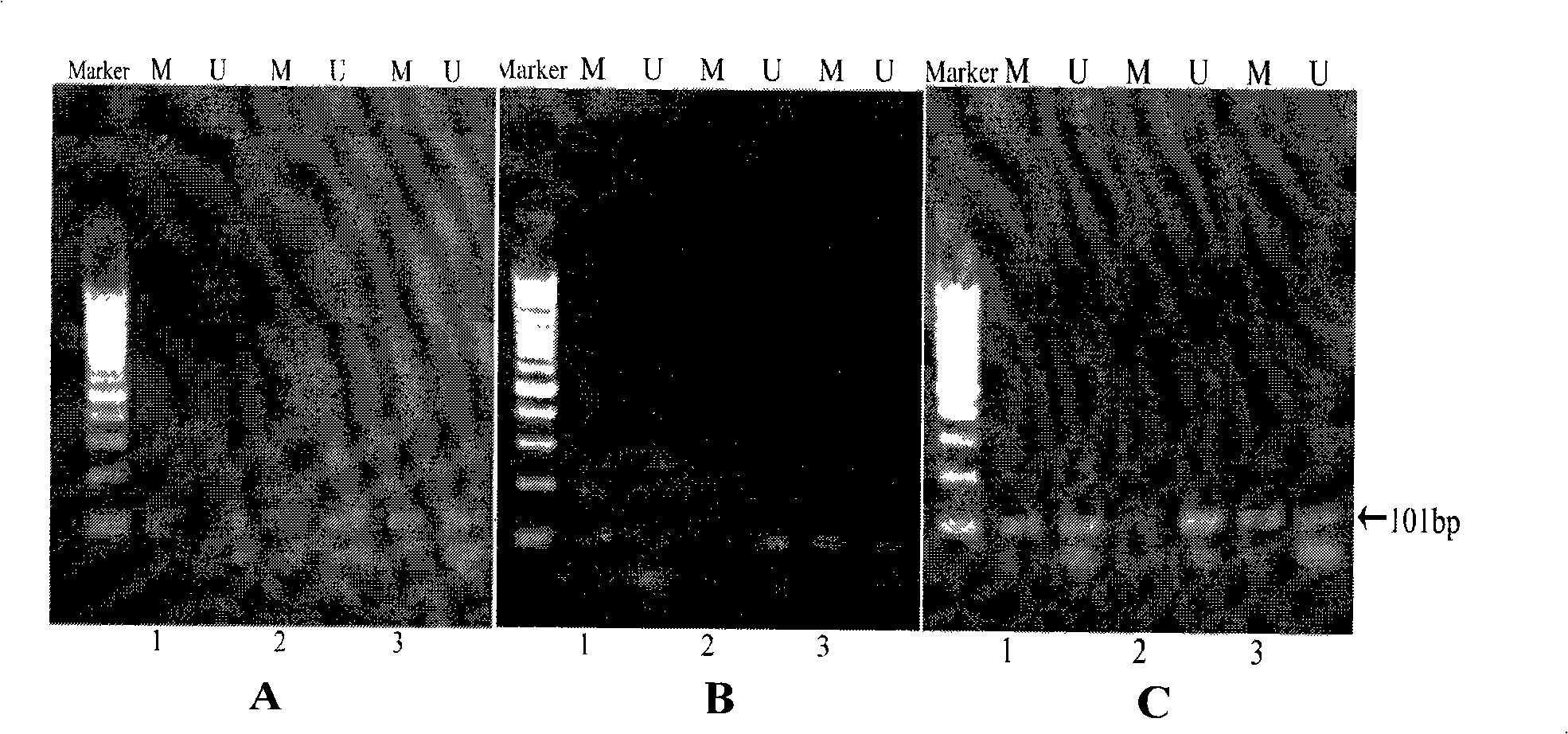
[0114] Embodiment 2: Comparative experiment of the method of the present invention and traditional method, kit method detection GSTP1 gene methylation
[0115] Design and synthesize methylation-specific primers and unmethylation-specific primers based on the GSTP1 gene, including forward primers and reverse primers (synthesized by Shanghai Bocai Biotechnology Co., Ltd.):
[0116] Methylated Forward Primer (MF): TTCGGGGTGTAGCGGTCGTC
[0117] Methylation Reverse Primer (MR): GCC CCAATACTAAATCACGACG
[0118] Amplified fragment length: 101bp
[0119] Unmethylated forward primer (UF): GATGTTTGGGGTGTAGTGGTTGTT
[0120] Unmethylated reverse primer (UR): CCACCCCAATACTAAATCACA ACA
[0121] Amplified fragment length: 101bp
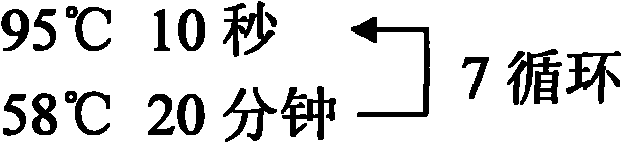
[0122] The PCR was divided into 2 tubes for amplification, one tube was amplified with methylation-specific primers, and the other tube was amplified with unmethylation-specific primers. The PCR reaction system and conditions were the same as in Example 1 (where...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com