Method for checking monocyte hyperplasia Listeria
A technology of mononucleosis and living bacteria, applied in the direction of biochemical equipment and methods, microbial measurement/inspection, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
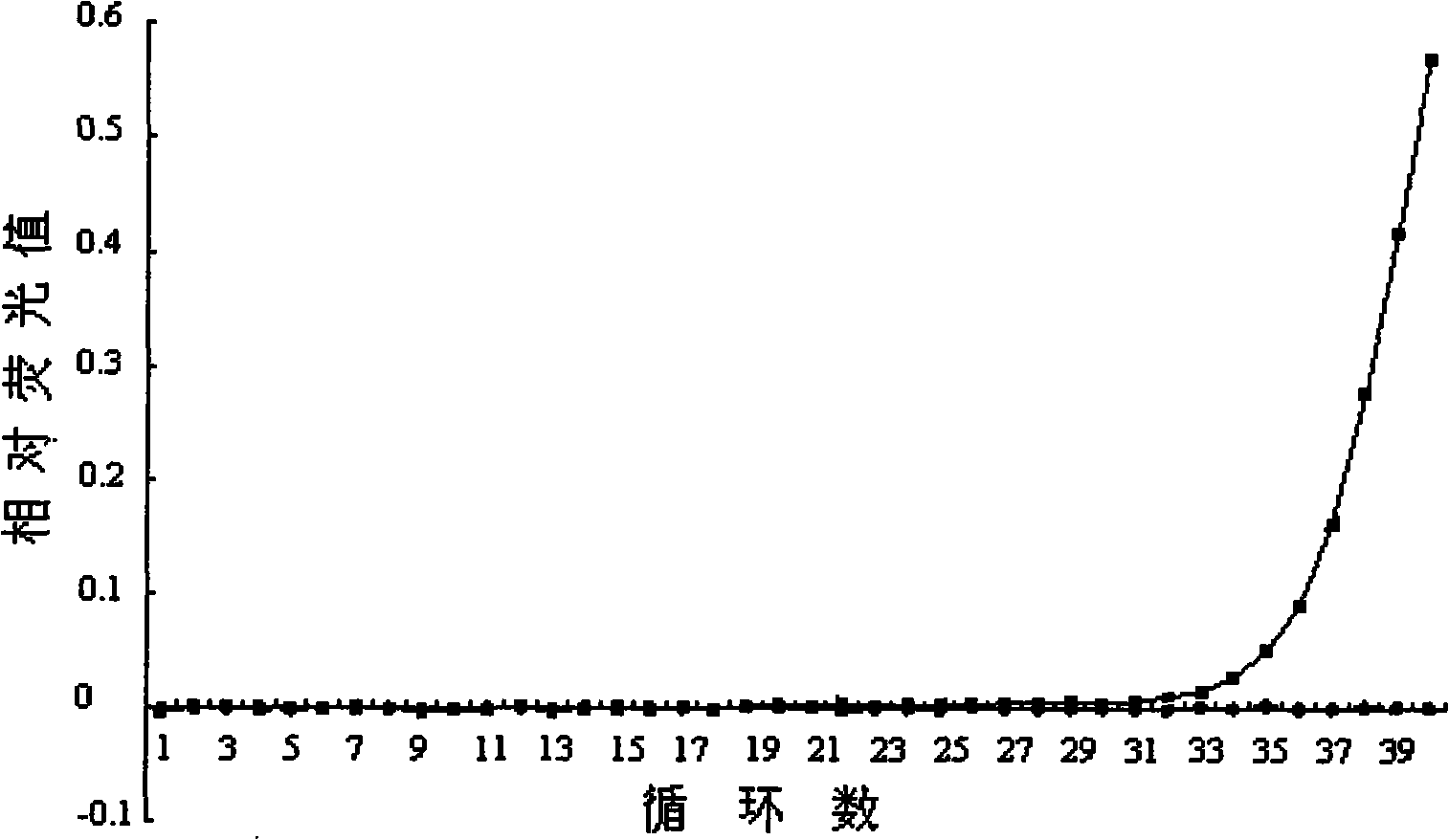
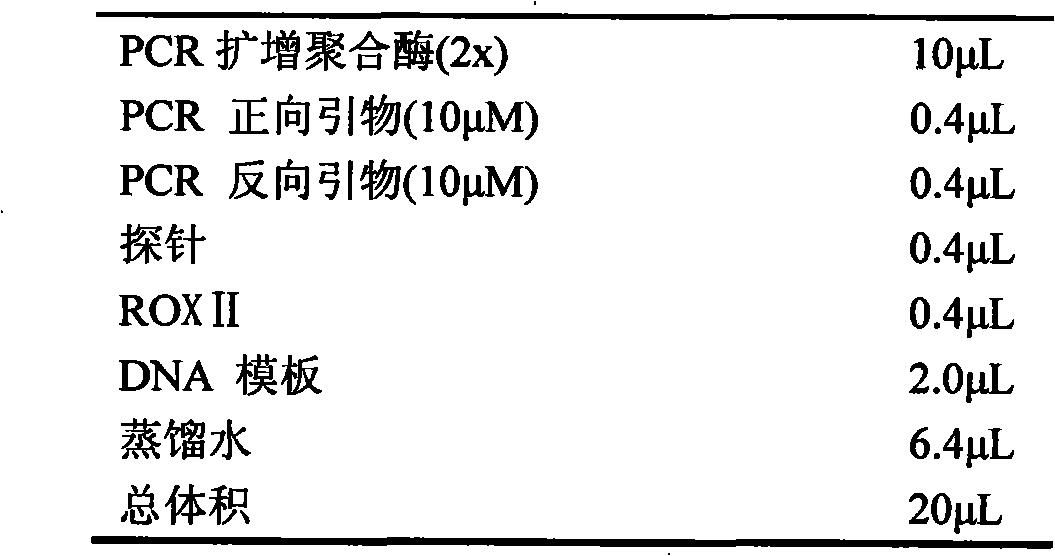
Image
Examples
specific Embodiment approach 1
[0020] Specific embodiment one: the method for the detection of Listeria monocytogenes viable bacteria in this embodiment is carried out according to the following steps:
[0021] 1. Take 25g of the article to be tested and place it in 225mL tryptone soybean yeast extract broth for 8 hours, then take 4mL of the culture solution and put it in a 5mL epoxy resin tube, and centrifuge at room temperature at a speed of 14000r / min 3min, get the fungus slime;
[0022] 2. Resuspend the obtained bacteria slime in a mixture of 3 mL of methanol, ether and ammonia, then resuspend in 1 mL of lysate, then add 0.2 g of glass beads with a diameter of 0.2 mm, and vortex at 2000 rpm 5min. Then centrifuge for 3min with the relative centrifugal force of 14000r / min;
[0023] 3. Take 700 μL of the suspension after centrifugation and mix it with the same volume of water-saturated phenol with a pH value of 5.5 in a 1.5 mL epoxy resin tube, incubate at 68°C for 5 minutes, and then centrifuge at room ...
specific Embodiment approach 2
[0035] Embodiment 2: The difference between this embodiment and Embodiment 1 is that the DEPC water in step 7 is MiliQ pure water treated with DEPC (diethypyrocarbonate, diethyl pyrocarbonate) and sterilized by high temperature and high pressure. Other steps and parameters are the same as those in Embodiment 1.
specific Embodiment approach 3
[0036] Embodiment 3: The difference between this embodiment and Embodiment 1 is: the volume ratio of methanol:ether:ammonia in the mixture of methanol, ether and ammonia in step 2 is 1:1:1. Other steps and parameters are the same as those in Embodiment 1.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com