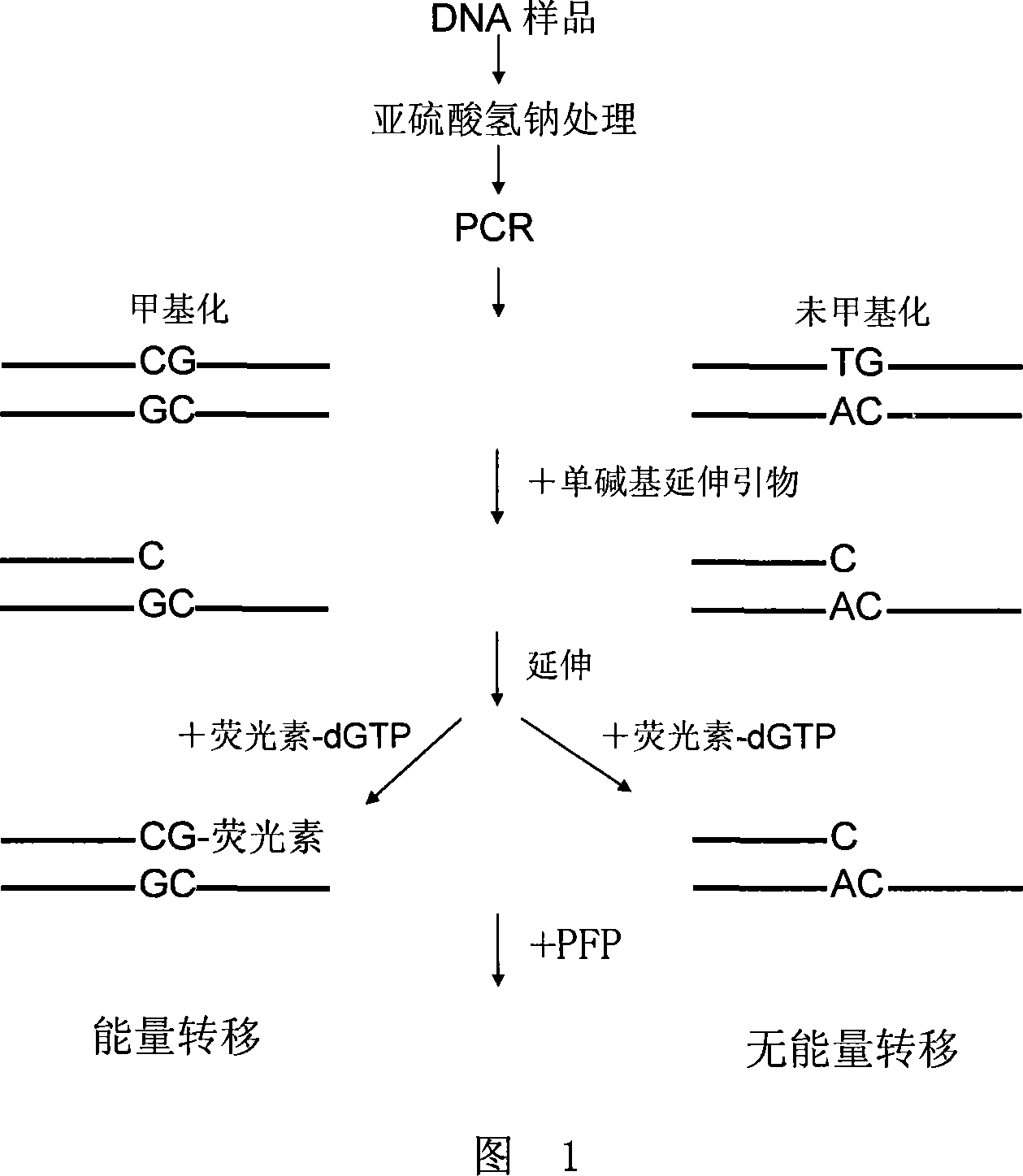
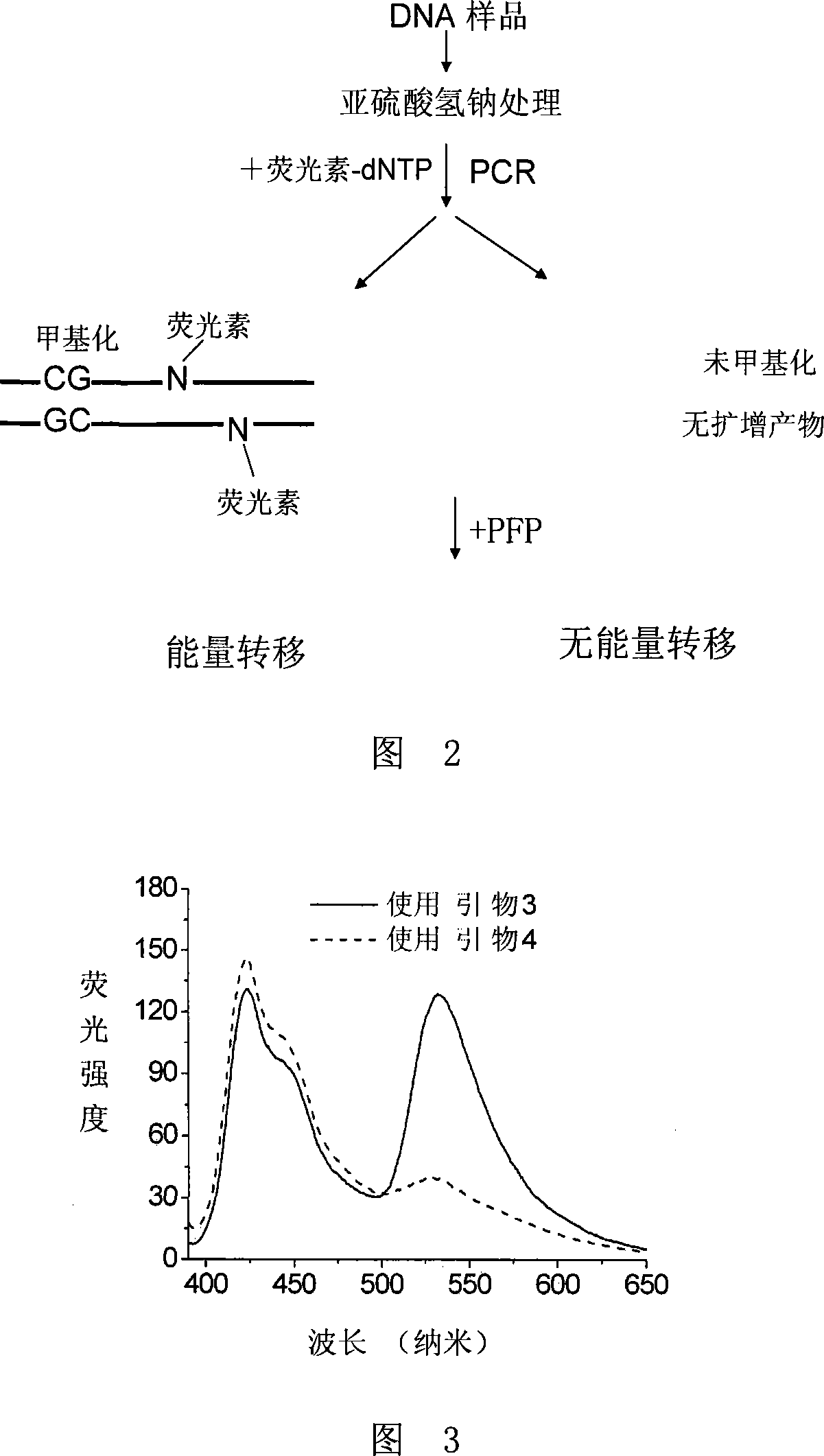
Method for detecting DNA methylation
A methylation and demethylation technology, applied in biochemical equipment and methods, microbiological determination/inspection, etc., can solve the problems of expensive equipment, low methylation level, limited sensitivity, etc., and reach the detection DNA concentration Low, sensitive detection, high sensitivity effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0062] Example 1, the application of PFP1 in the detection of plasmid 2 methylation
[0063] PFP1
[0064] Formula (II)
[0065] 1. Synthesis of PFP1
[0066] The structure of PFP1 is shown in formula (II), and the synthesis method is as follows:
[0067] Add 50mL of toluene, 5.2g of neopentyl glycol and 0.5g of p-toluenesulfonic acid to a 100mL single-necked bottle, reflux for 24 hours, evaporate the solvent and separate on a silica gel column (eluent: dichloromethane) to obtain 4.8g of 1,4- Neopentyldiboronate. 1 H NMR (400 MHz, CDCl 3 ): δ (ppm) 7.78 (4H, s), 3.77 (8H, s), 1.02 (12H, s).
[0068] Add 5.4mL toluene and 3.6mL 2M potassium carbonate to a 25mL two-necked flask, and after bubbling nitrogen gas for 30 minutes, add 325 mg of 2,7-dibromo-9,9-bis(6-bromohexyl)fluorene, 151 mg 1,4-Neopentyldiborate and 20 mg tetrakis(triphenylphosphine)palladium, stirred and reacted at 95°C under nitrogen for 24 hours, added chloro...
Embodiment 2
[0087] Example 2, Application of PFP1 in Plasmid 1 Methylation Detection
[0088] 1. Synthesis of PFP1
[0089] Same as Step 1 of Example 1.
[0090] Two, sodium bisulfite treatment
[0091] 500 ng of plasmid 1 was treated according to the literature to deaminate the unmethylated cytosine in the DNA to be tested into uracil, embed it in agarose, and store it at 4°C.
[0092] 3. PCR amplification
[0093] Same as Step 3 of Example 1.
[0094] 4. Single base extension reaction
[0095] Same as Step 4 of Example 1.
[0096] 5. Measurement of Fluorescence Spectroscopy
[0097] Same as Step 5 of Example 1.
[0098] Figure 4 shows the fluorescence spectra obtained by single-base extension with primer 3 and primer 4 respectively. When primer 4 is used, there is a significant energy transfer, but when primer 3 is used, only background signals appear, indicating that the target CpG site is demethylated. of.
Embodiment 3
[0099] Example 3, the application of PFP1 in the methylation detection of the mixture of plasmid 1 and plasmid 2
[0100] 1. Synthesis of PFP1
[0101] Same as Step 1 of Example 1.
[0102] Two, sodium bisulfite treatment
[0103] Plasmid 1 and plasmid 2 were mixed, and the mass ratio of plasmid 1 and plasmid 2 was 1:1. 500ng of the mixture was treated according to the literature to deaminate the unmethylated cytosine in the DNA to be tested into uracil, embed it in agarose, and store it at 4°C.
[0104] 3. PCR amplification
[0105] Same as Step 3 of Example 1.
[0106] 4. Single base extension reaction
[0107] Same as Step 4 of Example 1.
[0108] 5. Measurement of Fluorescence Spectroscopy
[0109] Same as Step 5 of Example 1.
[0110] Figure 5 shows the fluorescence spectra obtained by single-base extension with primers 3 and 4, respectively. Significant energy transfer occurred when primers 3 and 4 were used, indicating that the target CpG site was partially methyl...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com