Recombinant microorganism
一种重组微生物、微生物的技术,应用在微生物领域,能够解决收量降低等问题
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0065] Example 1 epr Construction of Plasmid for Gene Deletion
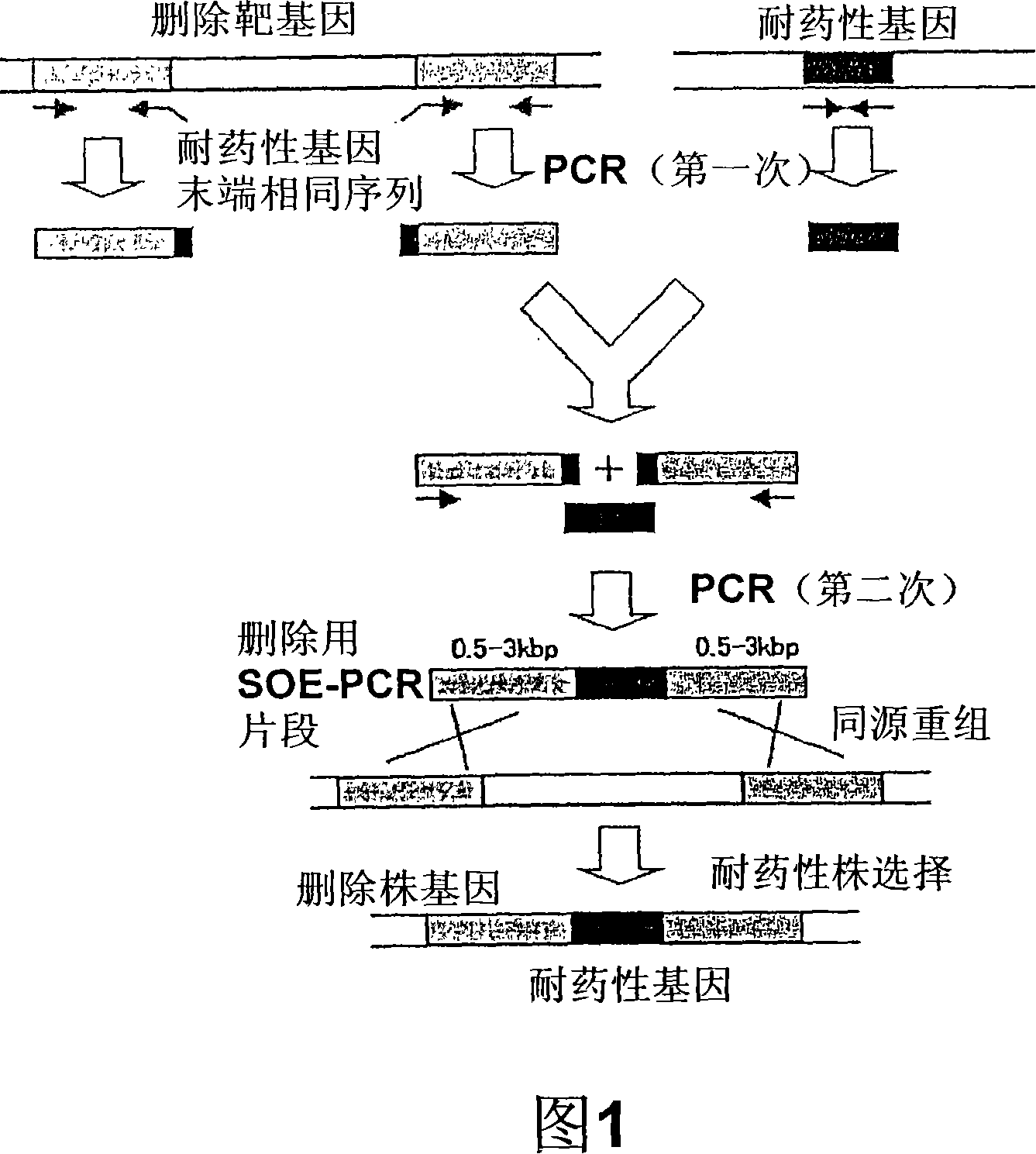
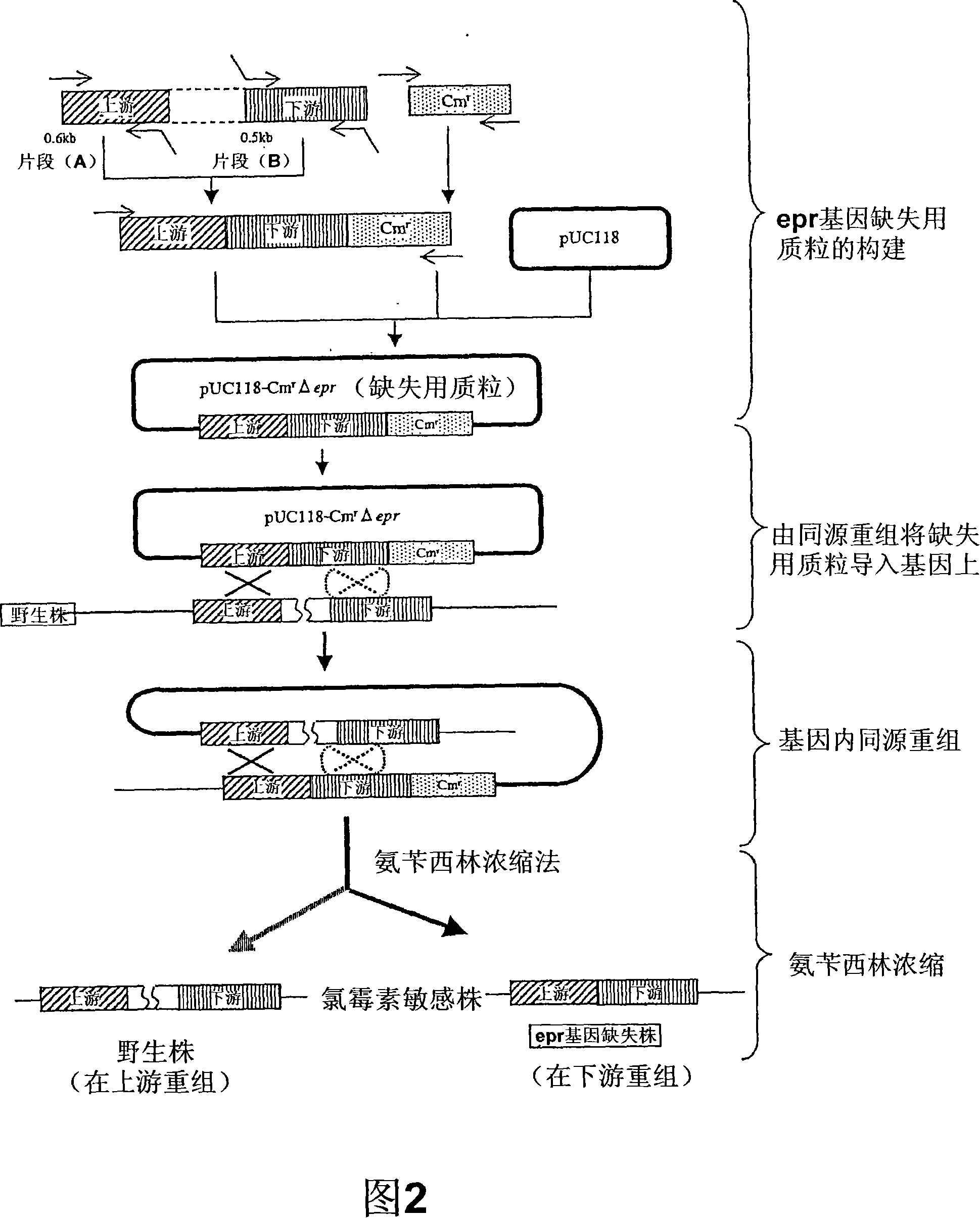
[0066] Using the genomic DNA extracted from 168 strains of Bacillus subtilis as a template, use the eprfw1 and eprUpr, as well as the primer sets of eprDNf and eprrv-rep shown in Table 2, respectively, to modulate the genomic DNA and epr The 0.6 kb fragment adjacent upstream of the gene (A) and the 0.5 kb fragment adjacent downstream (B). And prepared the upstream of the chloramphenicol resistance gene from other plasmid pC194 (J. Bacteriol. 150 (2), 815 (1982)) and the one from plasmid pUB110 (Plasmid 15, 93 (1986)) repU A 1.2 kb fragment (C) linked to the promoter region of the gene. Then, the obtained three fragments (A), (B), (C) were mixed, as a template, using the primers eprfw2 and Cmrv2 of Table 2 to perform SOE-PCR, and the sequence of (A) (B) (C) These three fragments were ligated to obtain a 2.2 kb DNA fragment (refer to Fig. 2). The end of the DNA fragment was smoothed and 5′-phosphorylated, and insert...
Embodiment 2
[0067] Example 2 Using plasmids for deletion epr Construction of gene deletion strain
[0068] Using the competent cell transformation method (J. Bacteriol. 93, 1925 (1967)), the epr The gene deletion plasmid pUC118-CmrΔepr was introduced into 168 strains of Bacillus subtilis and obtained in the epr A transformant that is fused with genomic DNA by homologous recombination of a single crossover between the upstream region of the gene or the region equivalent to the downstream region, and the transformant has a chloramphenicol resistance index. The obtained transformant was inoculated on LB medium, and after culturing at 37°C for two hours, the competent cell induction operation was performed again, thereby inducing repeated presence in the genome epr Intragenic homologous recombination between regions on genes or downstream regions. As shown in Figure 2, in the case of homologous recombination in a region different from the time of plasmid introduction, the chloramphenicol resi...
Embodiment 3
[0069] Example 3 Construction of 8-fold deletion strain of protease gene
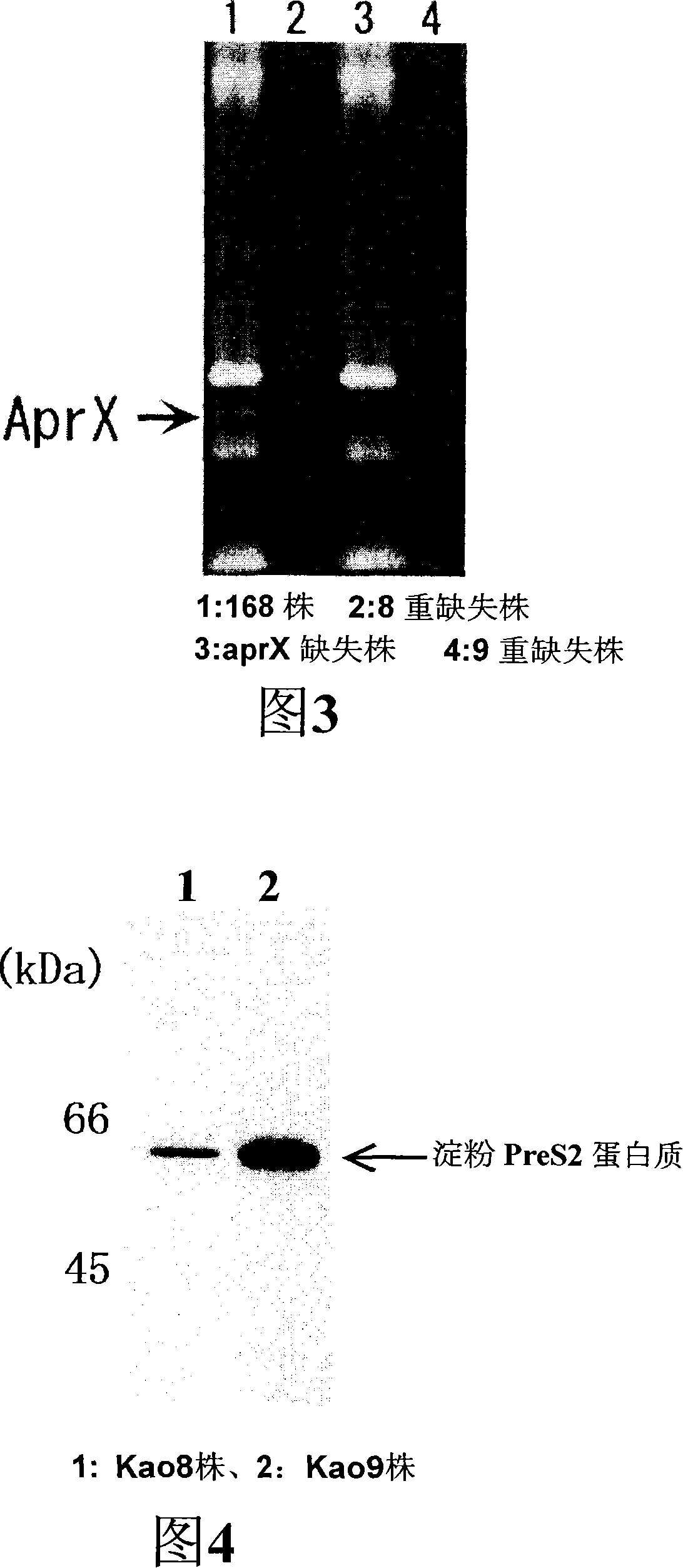
[0070] For epr Gene-deleted strains, carried out with epr The same gene deletion wprA The gene deletion serves as a further deletion. That is, as in Example 1, the structure wprA The plasmid pUC118-CmrΔwprA for gene deletion is obtained by introducing the constructed plasmid into genomic DNA and subsequent homologous recombination in the genome to obtain the deletion of the wprA gene epr Gene and wprA Double deletion of genes. After that, repeat the same operation, using mpr , nprB , bpr , nprE , vpr , aprE Each gene was deleted, and finally a protease 8-fold deletion strain with 8 protease genes deleted was constructed and named as Kao8 strain. The primer sequences used in each deletion are shown in Table 2, and the primers are the same as those shown in Example 1. epr The corresponding relationship of the primers used in the gene deletion is shown in Table 3.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com