Multiple PCR method based on selective probe and application thereof
A selectivity and probe technology, applied in multiplex PCR methods and its application fields, can solve problems such as amplification of multiple DNA sequences, difficult conditions to control, primer-primer dimer aggregation, etc., to enhance design sensitivity and simplicity Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0058] (1) Construction of selective probes
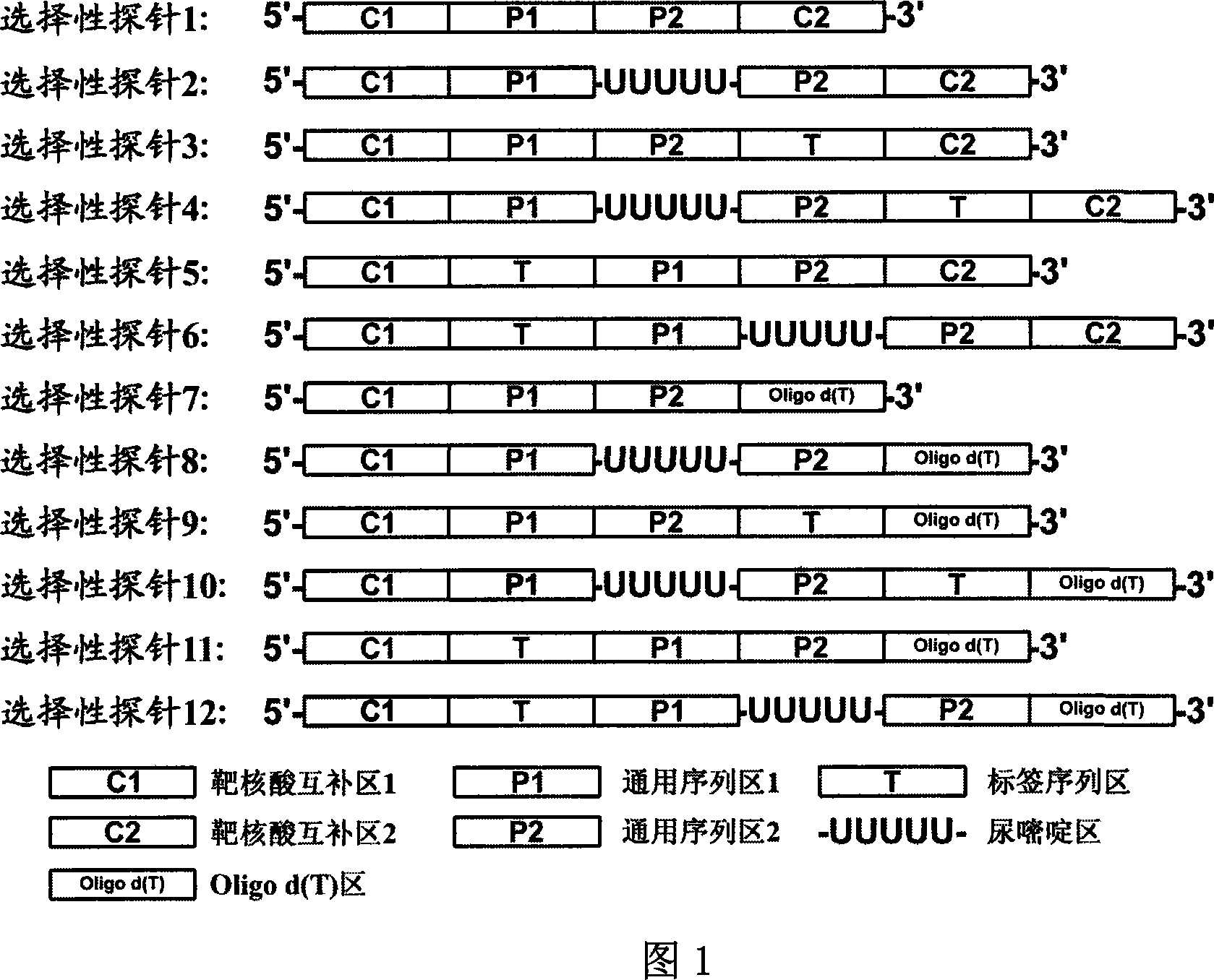
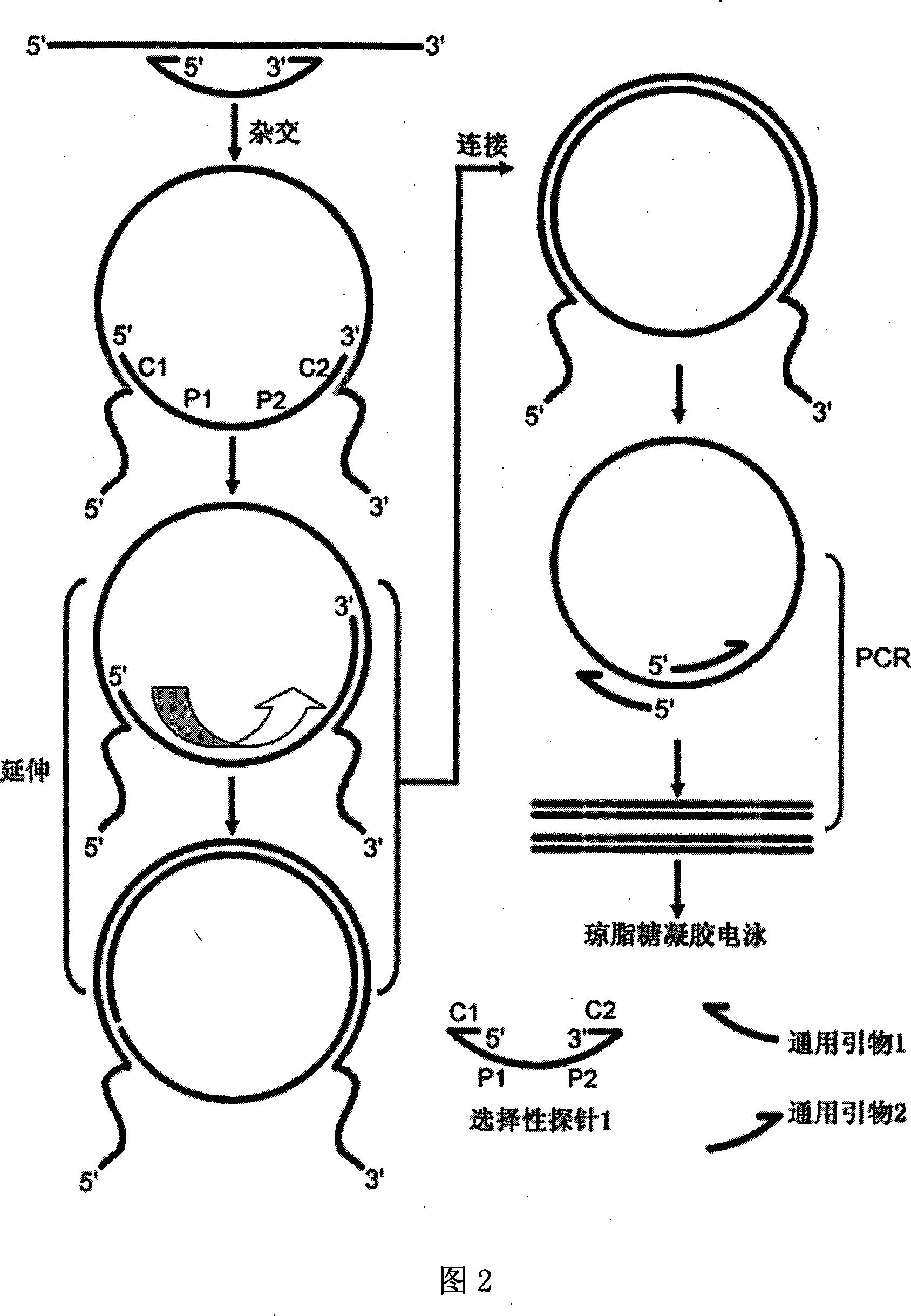
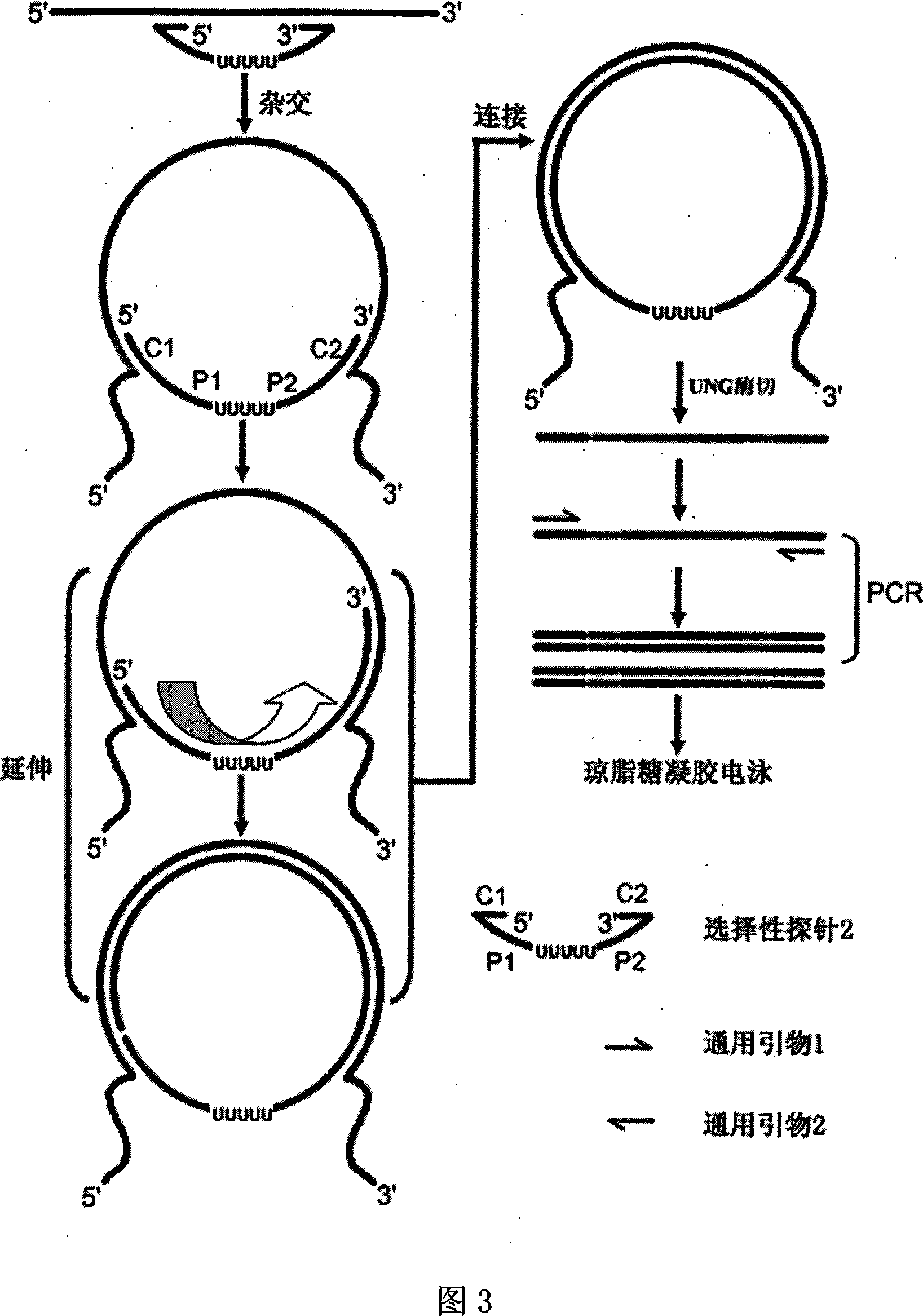
[0059] Fig. 1 is a schematic diagram of the structure of several selective probes of the present invention, as can be seen from Fig. 1, the selective probe provided by the present invention comprises at least four partitions, and it is characterized in that the middle part of the selective probe is two universal sequence regions ( P1 and P2 regions in Figure 1), the 5'- and 3'-ends of the selective probe are respectively the same nucleic acid single strand of the target double-stranded nucleic acid or the 5'- and 3'-ends of the target single-stranded nucleic acid. Two target nucleic acid complementary regions (C1, C2 districts in Fig. 1) that are complementary to each other, and, when the starting nucleic acid template is RNA or mRNA, the target nucleic acid complementary region of the 3'-end of the selective probe can be Olig d( T) (Oligo d(T) region in Figure 1), the length of Oligo d(T) is 8-40mer. There can also be a uracil re...
Embodiment 2
[0090] This example provides a multiplex PCR method for simultaneously detecting two CpG island sequences in the promoter region of the p16 gene.
[0091] (1) Design selective probes
[0092] Selective probe P16-V1 targets Chr9: 21,964,579-21,965,306 (728bp) CpG island sequence, its sequence is as follows:
[0093] 5′P-CATTCGCTAAGTGCTCGGAGCAGAATCGTCCAGTCGCAGT GAGG CATGTACCGTCGTTGT AGTCCTCCTTCCTTGCCAAC-3′
[0094] Wherein, the 5'- and 3'-end bold sequences are target nucleic acid complementary regions, and the 5'-terminal target nucleic acid complementary region is complementary to the 21965049-21965068 sequence of chromosome No. 9 (region C1 in Figure 1), and the 3'- The end-target nucleic acid complementary region is complementary to the sequence of 21965144-21965163 of chromosome 9 (region C2 in Figure 1); italicized (region P1 in Figure 1) and underlined sequence (region P2 in Figure 1) is a general sequence region . The 5'- and 3'-terminal bases of the selective prob...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com