Compounds that inhibit hsp90 protein-protein interactions with iap proteins
A protein interaction and compound technology, applied in the direction of apoptosis-related proteins, mammalian proteins, peptide/protein components, etc., can solve the problem of undetectable
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
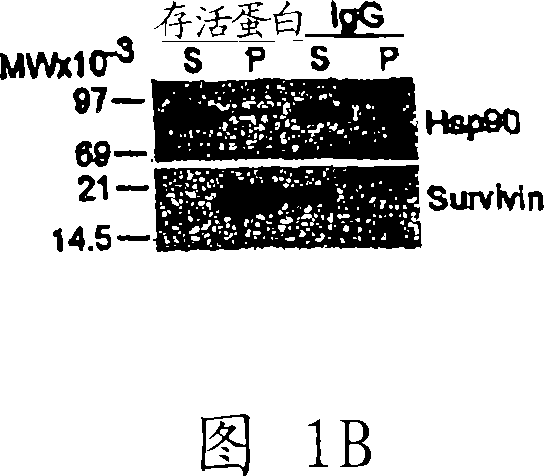
[0204] Example 1: Survivin interacts with Hsp90
[0205] Several experiments were performed to demonstrate the protein-protein interaction between survivin and Hsp90. Human cervical carcinoma HeLa cells and B lymphoma Raji cells were obtained from the American Type Culture Collection (ATCC) (Manassas, VA) and maintained in culture medium according to the supplier's instructions. For Western blotting, 12% sodium dodecyl sulfate (SDS) gels were transferred to nylon membranes and incubated with 1-5 μg of primary antibody. Rabbit polyclonal antibody against survivin was obtained from NOVUS Biologicals (Littleton, CO); monoclonal antibody (mAb) against Hsp90 was obtained from BD-Transduction Laboratories (catalogue # H38220; Lexington, KY); monoclonal anti-tubulin Antibodies were obtained from Sigma. Primary antibodies were developed with horseradish peroxidase (HRP)-conjugated secondary antibodies (Amersham, Piscataway, NJ) and a chemiluminescent kit (Amersham).
[0206] Figure...
Embodiment 2
[0209] Example 2: Proper folding of survivin is required for binding to Hsp90
[0210] Recombinant survivin (r-survivin) (10 mg / ml) or recombinant Hsp90 (r-Hsp90) (10 μg / ml) was mixed with bicarbonate buffer pH 9.5 (100 μl / well) in wells of a plastic microtiter plate (Immulon TM -2, Dynatech Laboratories, Chantilly, VA) at 4° C. for 18 hours for ELISA experiments. Bound proteins were blocked with 3% gelatin at 37°C for 1 hour, washed with washing buffer (TBS pH 7.4, 0.1% Tween TM , 0.1% BSA), and then incubated with different concentrations of the test protein at 37° C. for 1 hour to determine whether the test protein binds to the solid-phase protein.
[0211] GST-Hsp90 (amino acids 1-732), recombinant GST-survivin, GST-survivin (C84A) were expressed in E. coli and bound to glutathione beads (Sigma). Hsp90α was cloned into pGex-4T3 (Amersham Pharmacia Biotech.) by PCR. Purified r-survivin lacking the GST box was treated with thrombin (Sigma, 20 U / ml in 50 mM Tris TM pH 7....
Embodiment 3
[0215] Example 3: Survivin binds to the N-terminus of Hsp90
[0216] For full-length Hsp90α (SEQ ID NO: 21; amino acids 1-732) or three fragments: N-Hsp90 (amino acids 1-272 of SEQ ID NO: 21), M-Hsp90 (amino acids 273- 617), and C-Hsp90 (amino acids 629-732 of SEQ ID NO: 21), the nucleotide sequences encoding them were cloned into pGex-4T3 (Pharmacia Biotech.) and pFLAG-CMV 6c by PCR using the BamHI cloning site (Sigma). GST fusions of full-length Hsp90 or individual Hsp90 fragments were expressed in E. coli and bound to glutathione beads (Sigma). Purified recombinant survivin lacking the GST box was obtained as described in Example 2.
[0217] Figure 3A shows the results of the survivin / Hsp90 pull-down experiment in vitro. Increasing concentrations of r-survivin (30, 100 and 300ng / 50μl reaction) were mixed with Sepharose TM - GST-Hsp90 (10 μg / 50 μl reaction) or incubated with GST fused to N-Hsp90, M-Hsp90 or C-Hsp90 region. The reaction was centrifuged to make Sepharose ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com