Assay for the diagnosis of dermatophytosis
a technology for dermatophytosis and assays, applied in the field of assays for the diagnosis of dermatophytosis, can solve the problems of long-term and often expensive systemic treatment, false negative results, and low sensitivity of techniques, and achieves the problem of not allowing the rapid and reliable distinction of closely related strains from the genus i>trichophyton
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Design of Primers and Probes
[0083]For the identification of T. tonsurans in a sample, the metalloprotease gene was chosen as a useful target region. Based on the comparison of oligonucleotide sequences, a set of primers (forward primer 5′ cy3-GGGAGGGAGACTAGTTG 3′ (SEQ ID NO: 10), reverse primer 5′ cy3-AATTTTTCGCCGCCAAG 3′(SEQ ID NO: 12)) and a species-specific probe (5′ C6-Amino-linker-GATCCTTGGCGTACGGATGCATA 3′(SEQ ID NO: 14)) for the detection of Trichophyton tonsurans were designed. The designed probes were checked for internal repeats, secondary structure, melting temperature and GC content.
[0084]The designed probe for T. tonsurans and some controls were spotted with the sciFLEXARRAYER S11 (Scienion AG, Germany) on a solid carrier material as microscopically small spots located at defined positions.
DNA Extraction from Dermatophyte Cultures
[0085]Cultures were performed on dermatophyte test medium agar (SIFIN, Germany, Tenn. 2102). The DNA from cultured T. tonsurans ...
example 2
Material and Methods
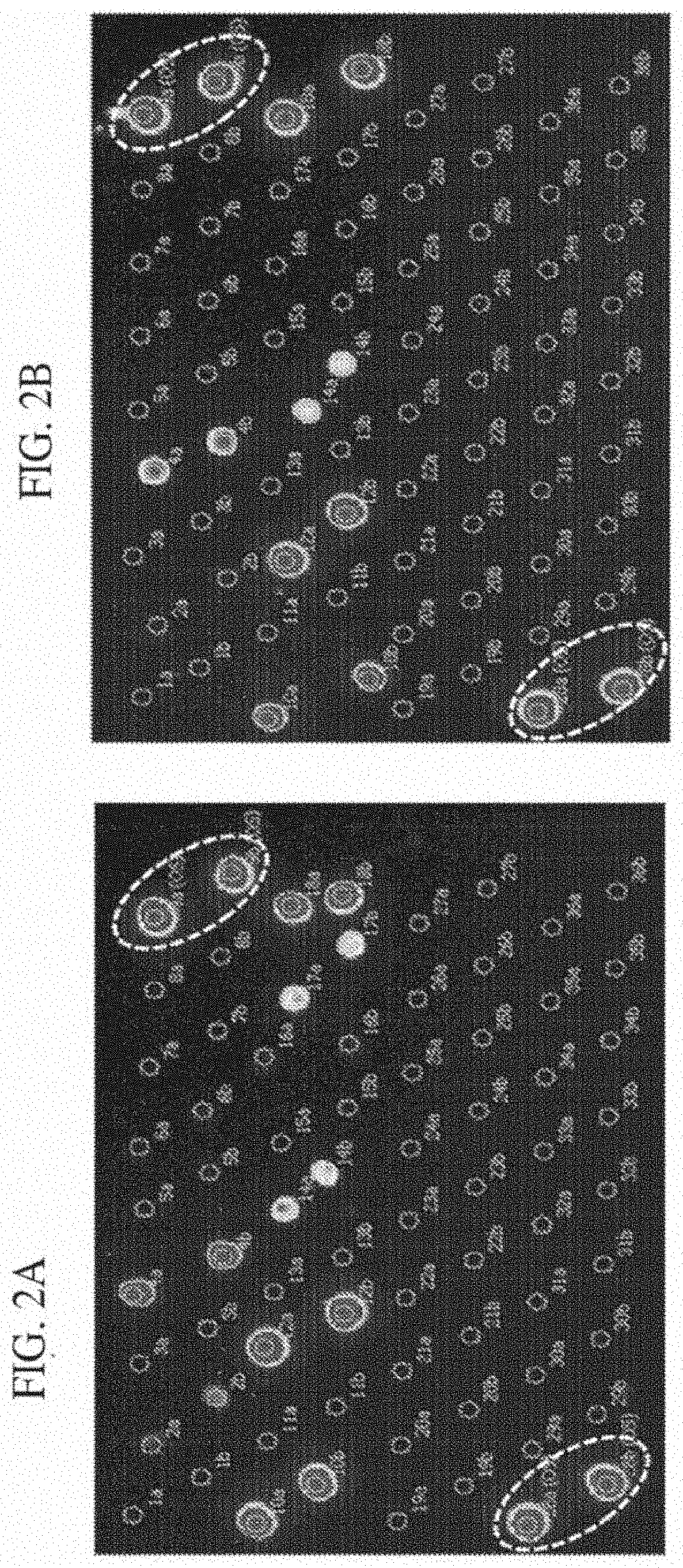
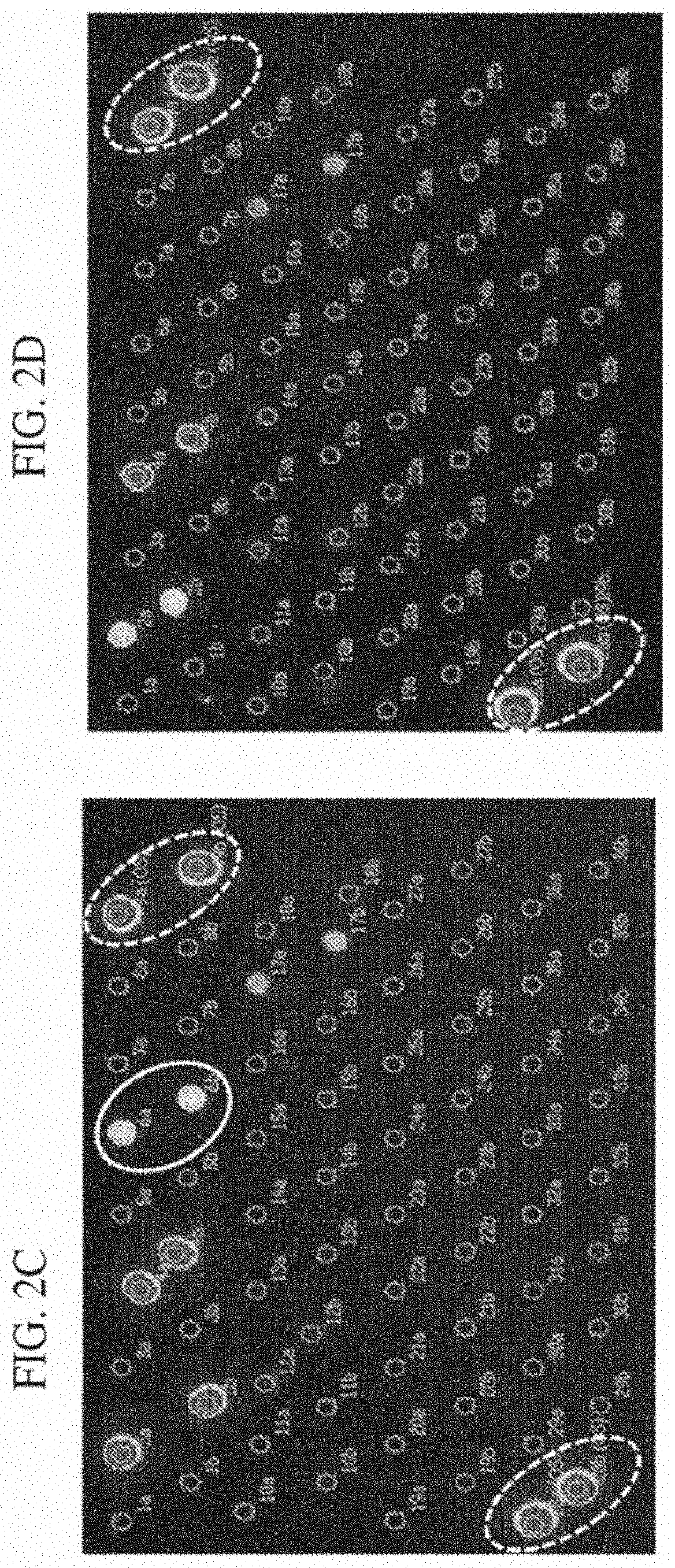
[0091]DNA microarrays consist of DNA molecules (probes) that differ from one another by their DNA sequence. When the DNA of an organism contains segments that match to these defined probes at the microarray, the complementary DNA regions bind together (hybridize). Due to fluorescence labeled primers that are used in the polymerase chain reaction (PCR), a positive hybridization between probe and amplified target sequence can be detected via microarray scanner. An evaluated positive signal means that the target sequence could be detected. In this example the detection of the dermatophyte Trichophyton benhamiae (yellow) and Trichophyton concentricum via DNA microarray will be shown. For the verification of probe specificity the most closely related species Trichophyton benhamiae (white) and Trichophyton benhamiae (african) were also included in the analysis. The used method based on the EUROIMMUN DNA microarray platform.
Design of Primers and Probes
[0092]For the iden...
example 3
Material and Methods
[0098]DNA microarrays consist of DNA molecules (probes) that differ from one another by their DNA sequence. When the DNA of an organism contains segments that match to these defined probes at the microarray, the complementary DNA regions bind together (hybridize). Due to fluorescence labeled primers that are used in the polymerase chain reaction (PCR), a positive hybridization between probe and amplified target sequence can be detected via microarray scanner. An evaluated positive signal means that the target sequence could be detected. In this example the detection of the dermatophyte Trichophyton benhamiae (yellow) and Trichophyton benhamiae (white / african) via DNA microarray will be shown. The used method based on the EUROIMMUN DNA microarray platform.
Design of Primers and Probes
[0099]For the identification of Trichophyton benhamiae (yellow) or Trichophyton benhamiae (white / african) in a sample the metalloprotease gene was chosen as a useful target region. Bas...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Fluorescence | aaaaa | aaaaa |
| Chemiluminescence | aaaaa | aaaaa |
| Radioactivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com