Quantitative assessment of biological impact using overlap methods
a biological network and overlap technology, applied in the field of quantitative assessment of biological impact using overlap methods, to achieve the effects of increasing the specificity of hyps, reducing cross-network redundancy, and increasing the abundance of activation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0021]As used herein, the following terms have the following definitions:
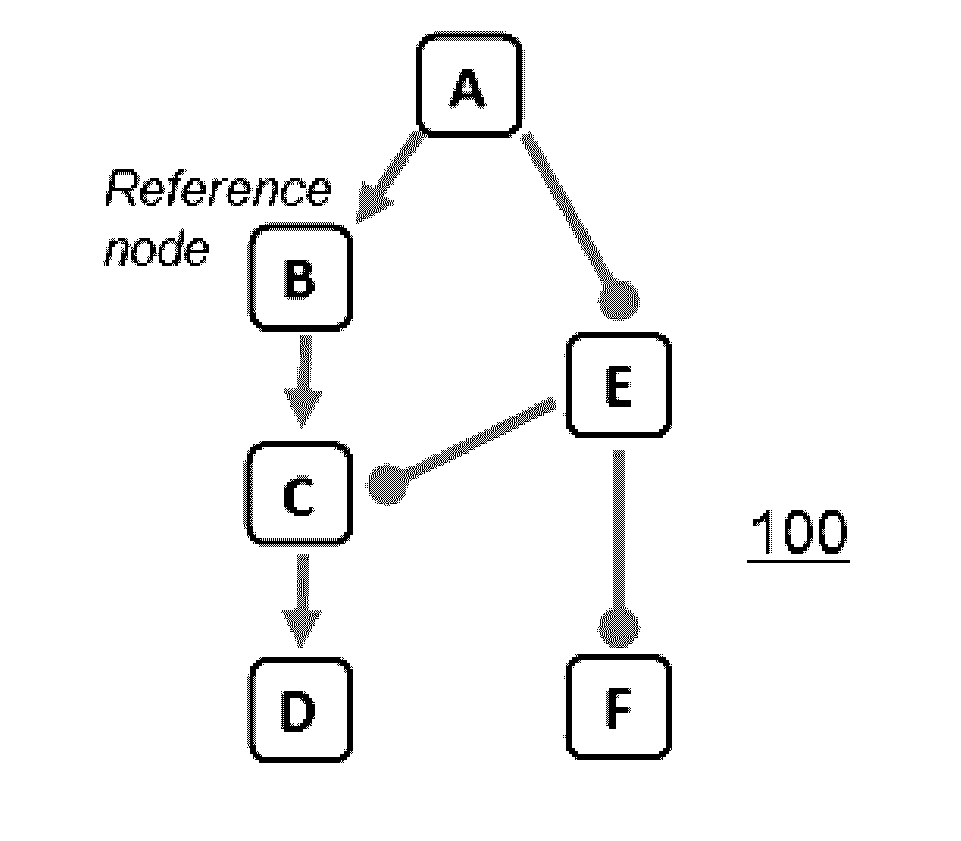
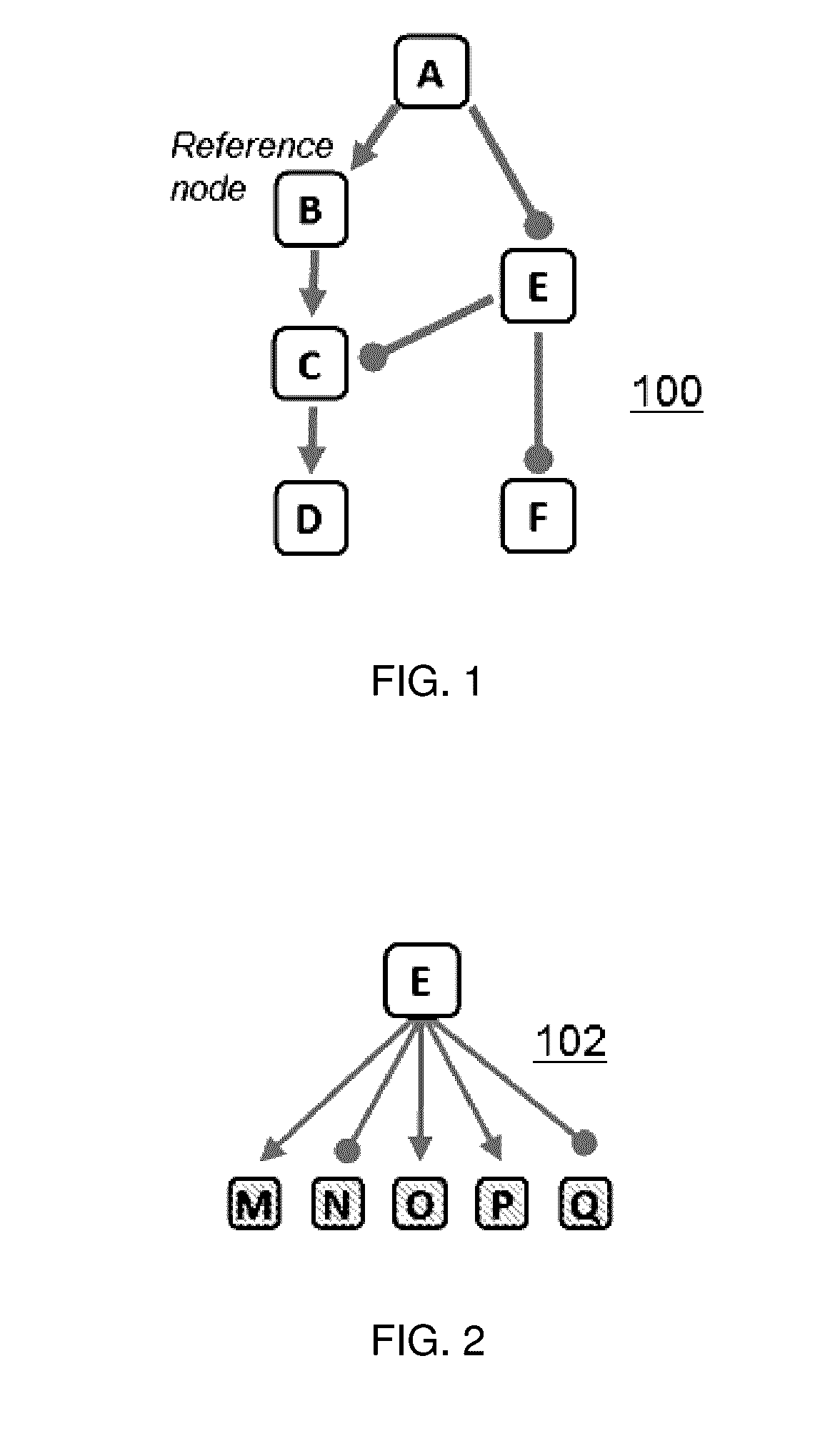
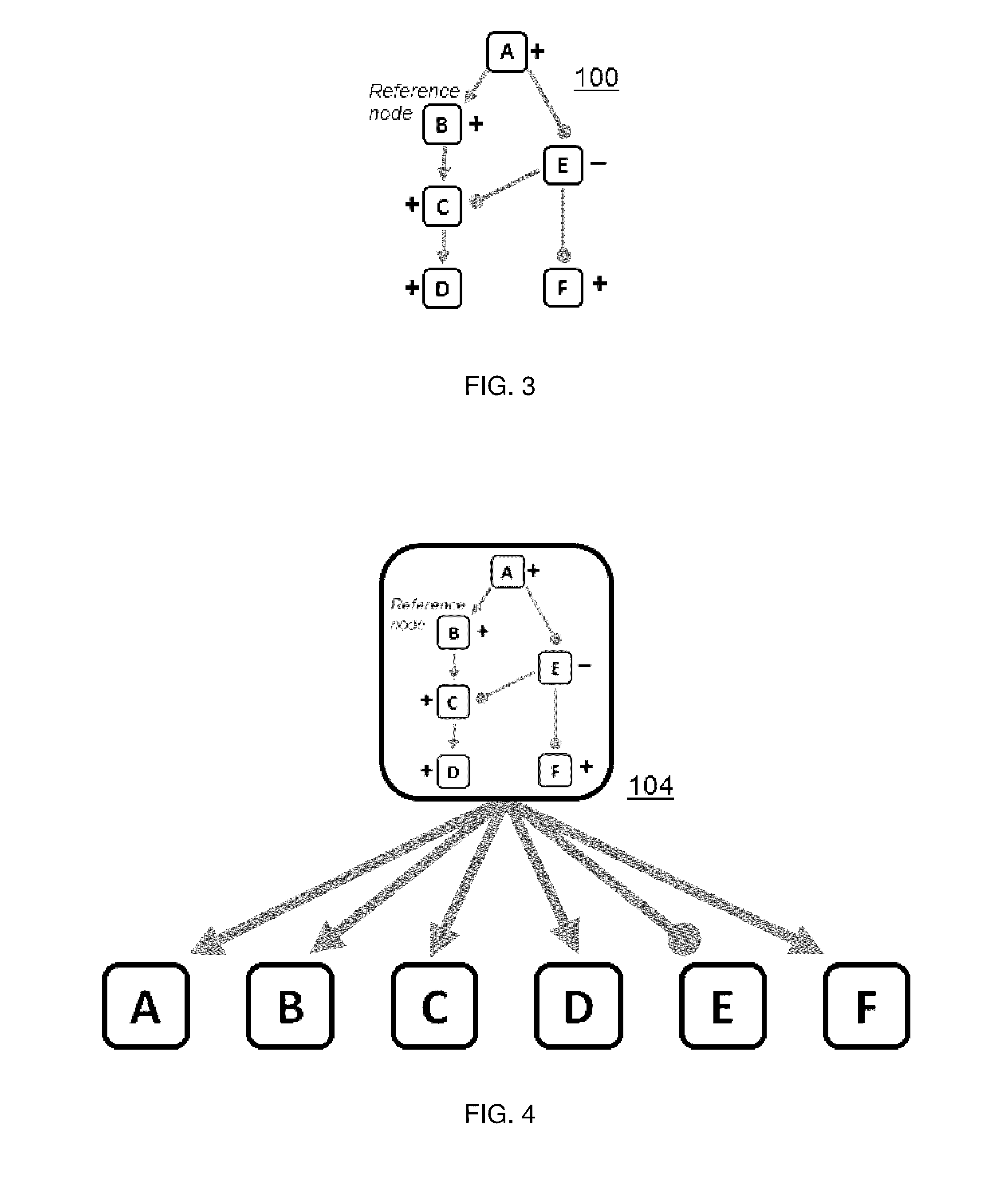
[0022]A “knowledge base” is a directed network, preferably of experimentally-observed casual relationships among biological entities and processes;
[0023]A “node” is a measurable entity or process;
[0024]A “measurement node” is a measured entity or process;
[0025]A “reference node” represents a potential perturbation to a node;
[0026]A “signature” is a collection of measurable node entities and their expected directions of change with respect to a reference node;
[0027]A “differential data set” is a data set that has data associated with a first condition, and data associated with a second condition distinct from the first condition;
[0028]A “fold change” is a number describing how much a quantity changes going from an initial to a final value, and is specifically computed by dividing the final value by the initial value;
[0029]A “signed graph” (i.e., a graph with a signed edge) is a representational structure that, i...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com