Methods and products related to genotyping and DNA analysis
a genotyping and dna technology, applied in the field of methods and products related to genotyping, can solve the problems of high cost, difficult identification of complex gene mutations contributing to diabetes disorders, and high cost of southern blotting, and achieve the effect of high throughpu
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Identification and Isolation of SNPs
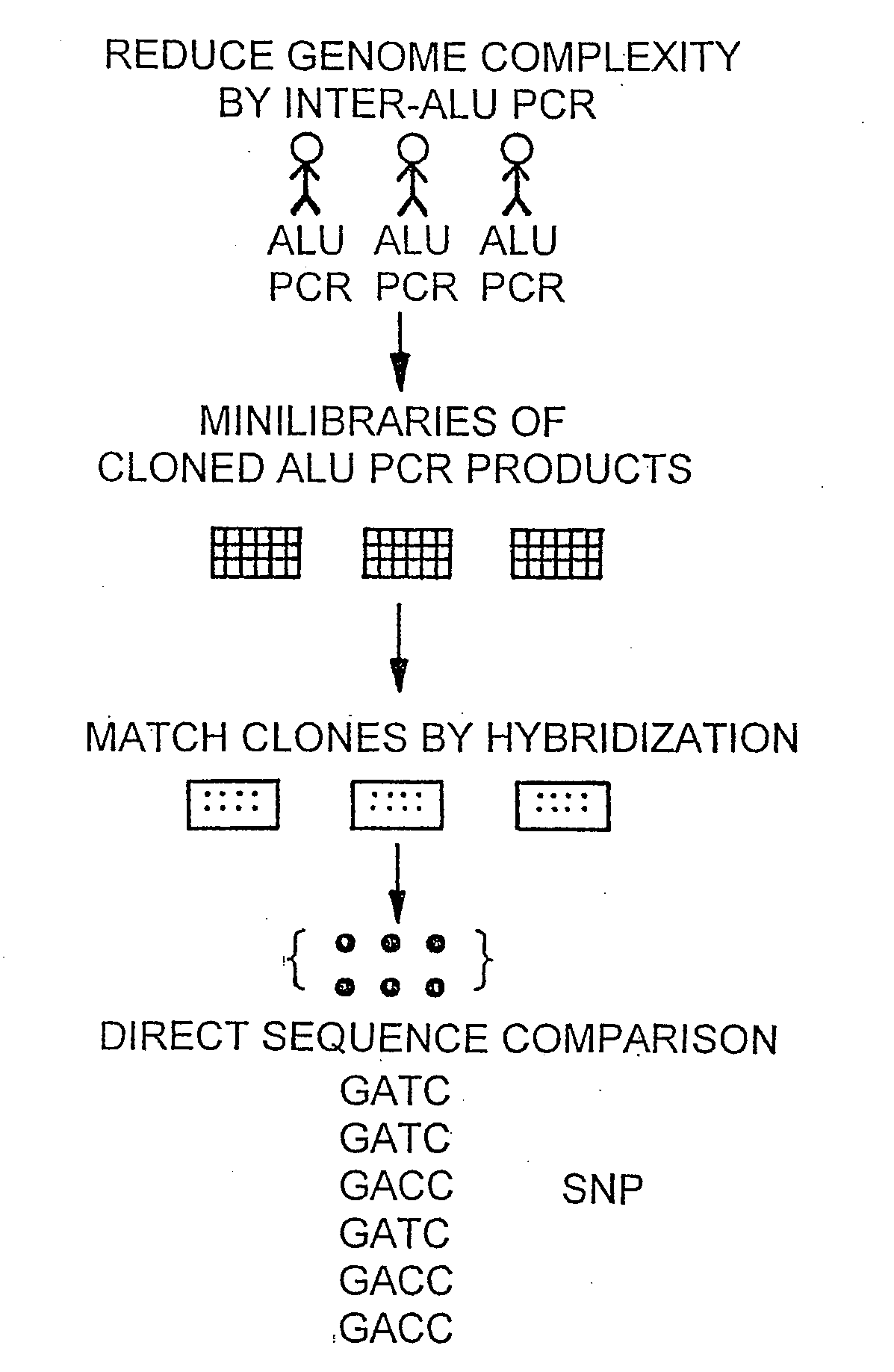
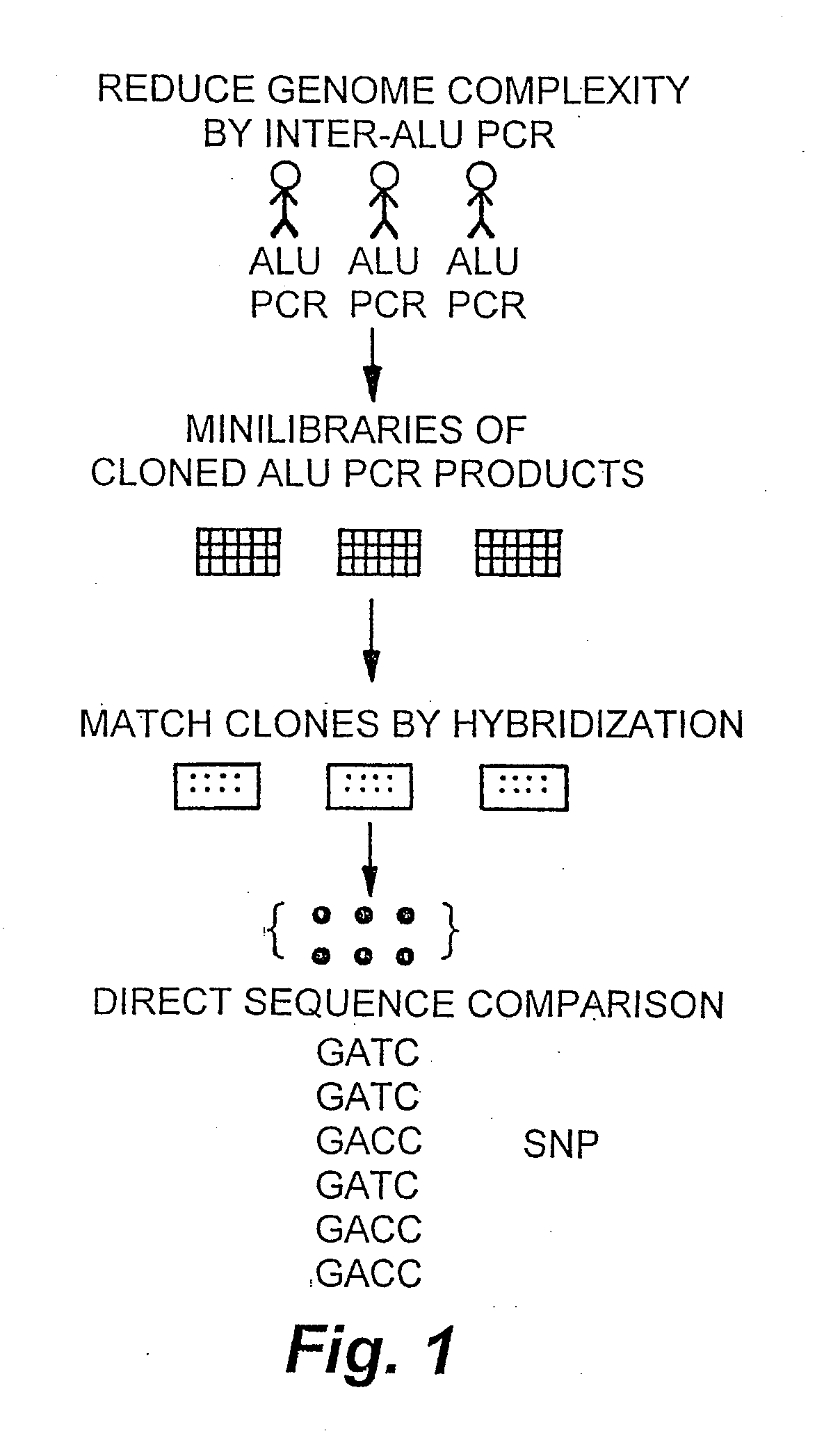
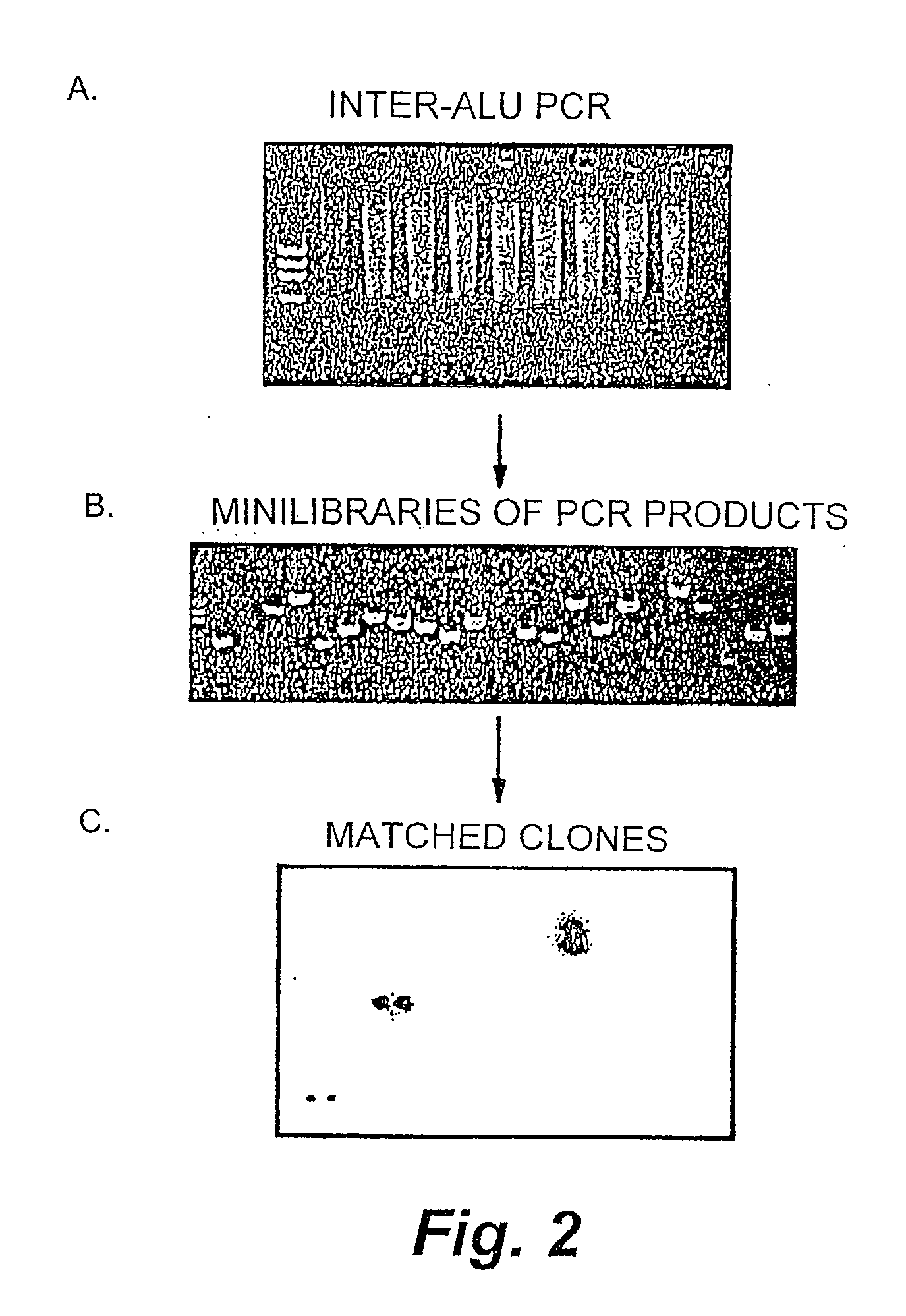
[0191]High allele frequency SNPs are estimated to occur in the human genome once every kilobase or less (Cooper et al., 1985). A method for identifying these SNPs is illustrated in FIG. 1. As shown in FIG. 1, inter-Alu PCR was performed on genomes isolated from three unrelated individuals. The PCR products were cloned, and a mini library was made for each of the 3 individuals. The library clone inserts were PCR-amplified and spotted on nylon filters. Clones were matched by hybridization into two sets of identical clones from each individual, for a total of 6 clones per matched clone set. These sets of clones were sequenced, and the sequences were compared in order to identify SNPs. This method of identifying SNPs has several advantages over the prior art PCR amplification methods. For instance, a higher quality sequence is obtained from cloned DNA than is obtained from cycle sequencing of PCR products. Additionally, every sequence represents a spe...
example 2
Allele-Specific Oligonucleotide Hybridization to Alu PCR SNPs
Methods
[0200]Inter-Alu PCR was performed using genomic DNA obtained from 136 members of 8 CEPH families (numbers 102, 884, 1331, 1332, 1347, 1362, 1413, and 1416) using the 8C Alu primer, as described above. The products from these reactions were denatured by alkali treatment (10-fold addition of 0.5 M NaOH, 2.0 M NaCl, 25 mM EDTA) and dot blotted onto multiple Hybond™ N+ filters (Amersham) using a 96-well dot blot apparatus (Schleicher and Schull). For each SNP, a set of two allele-specific oligonucleotides consisting of two 17-residue oligonucleotides centered on the polymorphic nucleotide residue were synthesized. Each filter was hybridized with 1 picomole 32P-kinase labeled allele-specific oligonucleotides and a 50-fold excess of non-labeled competitor oligonucleotide complementary to the opposite allele (Shuber et al., 1993). Hybridizations were carried out overnight at 52° C. in 10 mL TMAC buffer 3.0 M TMAC, 0.6% SDS...
example 3
Confirmation of SNP Identity
[0204]Allele-specific oligonucleotides are synthesized based on standard protocols (Shuber et al., 1997). Briefly, polynucleotides of 17 bases centering on the polymorphic site are synthesized for each allele of a SNP. Hybridization with DNA dots of IRS or DOP-PCR products affixed to a membrane were performed, followed by hybridization to end labeled allele-specific oligonucleotides under TMAC buffer conditions. These conditions are known to equalize the contribution of AT and GC base pairs to melting temperature, thereby providing a uniform temperature for hybridization of allele-specific oligonucleotides independent of nucleotide composition.
[0205]Using this methodology, genotypes of CEPH progenitors and their offspring are determined. The Mendelian segregation of each SNP marker confirms its identity as a SNP marker and accrued estimate of its relative allele frequency, hence, its likely usefulness as a genetic marker. Markers which yield complex segre...
PUM
| Property | Measurement | Unit |
|---|---|---|
| frequency | aaaaa | aaaaa |
| restriction fragment length polymorphism | aaaaa | aaaaa |
| simple sequence length polymorphisms | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com