In vivo screen using chemical inducers of dimerization
a dimerization and chemical inducer technology, applied in the field of enzyme evolution and drug screening in vivo, can solve the problems of limited to existing reactions, difficult protein engineering, and naive notions of how enzymes work
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
[0185]We have shown that Dex-Mtx can dimerize a LexA-DHFR and a B42-rGR protein chimera in vivo (Table I). (Lin, 2000) Dex-Mtx was assayed using both plate and liquid assays at extracellular concentrations of 1-100 μM. No activation was observed at concentrations ≦0.1 μM. 100 μM is the limit of Dex-Mtx solubility. Control experiments established that lacZ transcription is dependent on Dex-Mtx. There are only background levels of lacZ transcription when Dex-Mtx is omitted, LexA-DHFR is replaced with LexA, or B42-GR is replaced with B42. Likewise, a 10-fold excess of Mtx competes out Dex-Mtx-dependent LacZ transcription. Interestingly, of the 10 protein chimera combinations tested, Dex-Mtx could only activate lacZ transcription in the context of the LexA-eDHFR and the B42-(Gly6)-rGR chimeras (Table 1). None of the 9 other protein combinations tested worked. This result is consistent with our view that the Dex-Mtx systems (and other dimerization systems) could be further improved both ...
example 2
Cephalosporin Hydrolysis by the 908R Cephalosporinase in the Yeast Three-Hybrid System
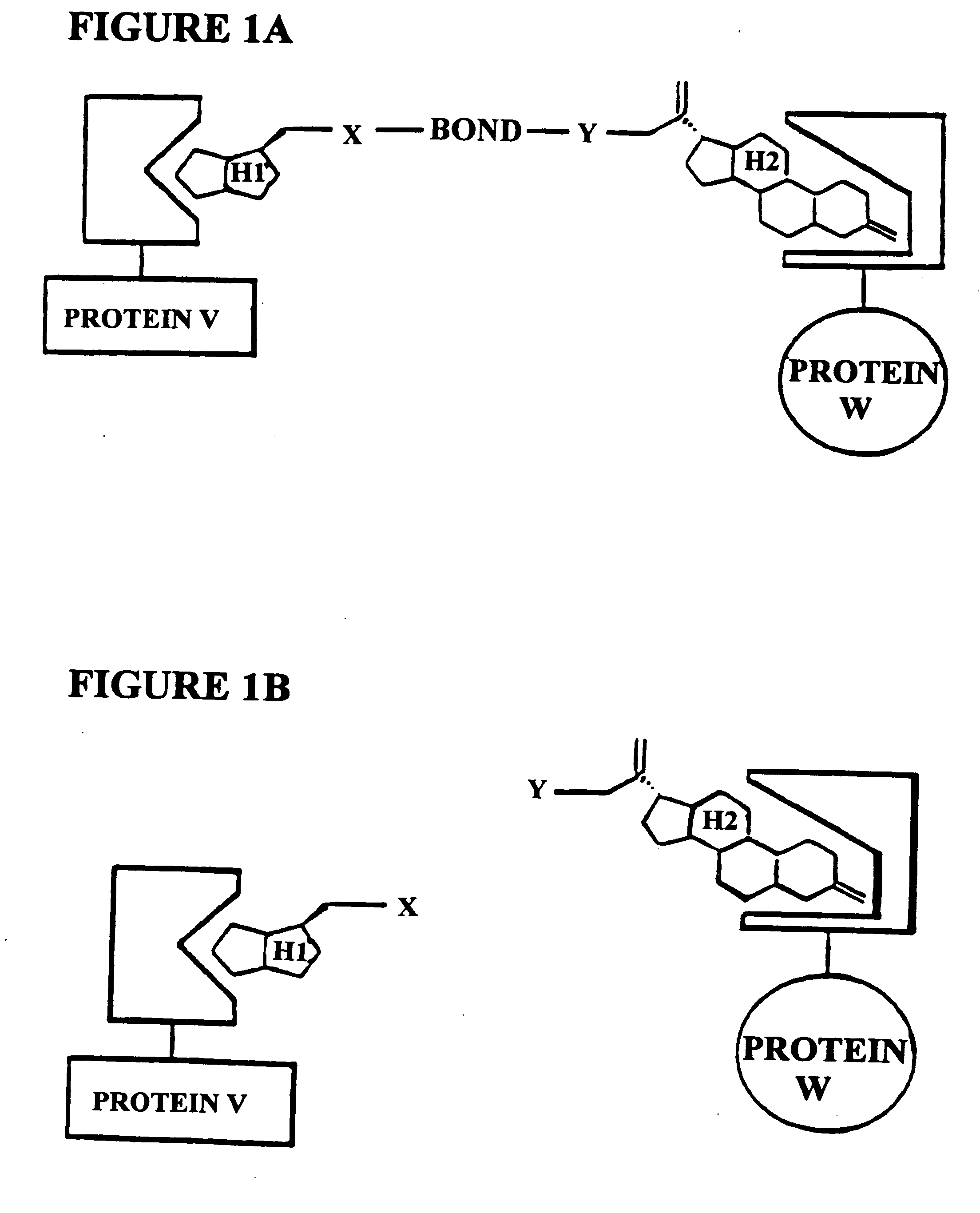
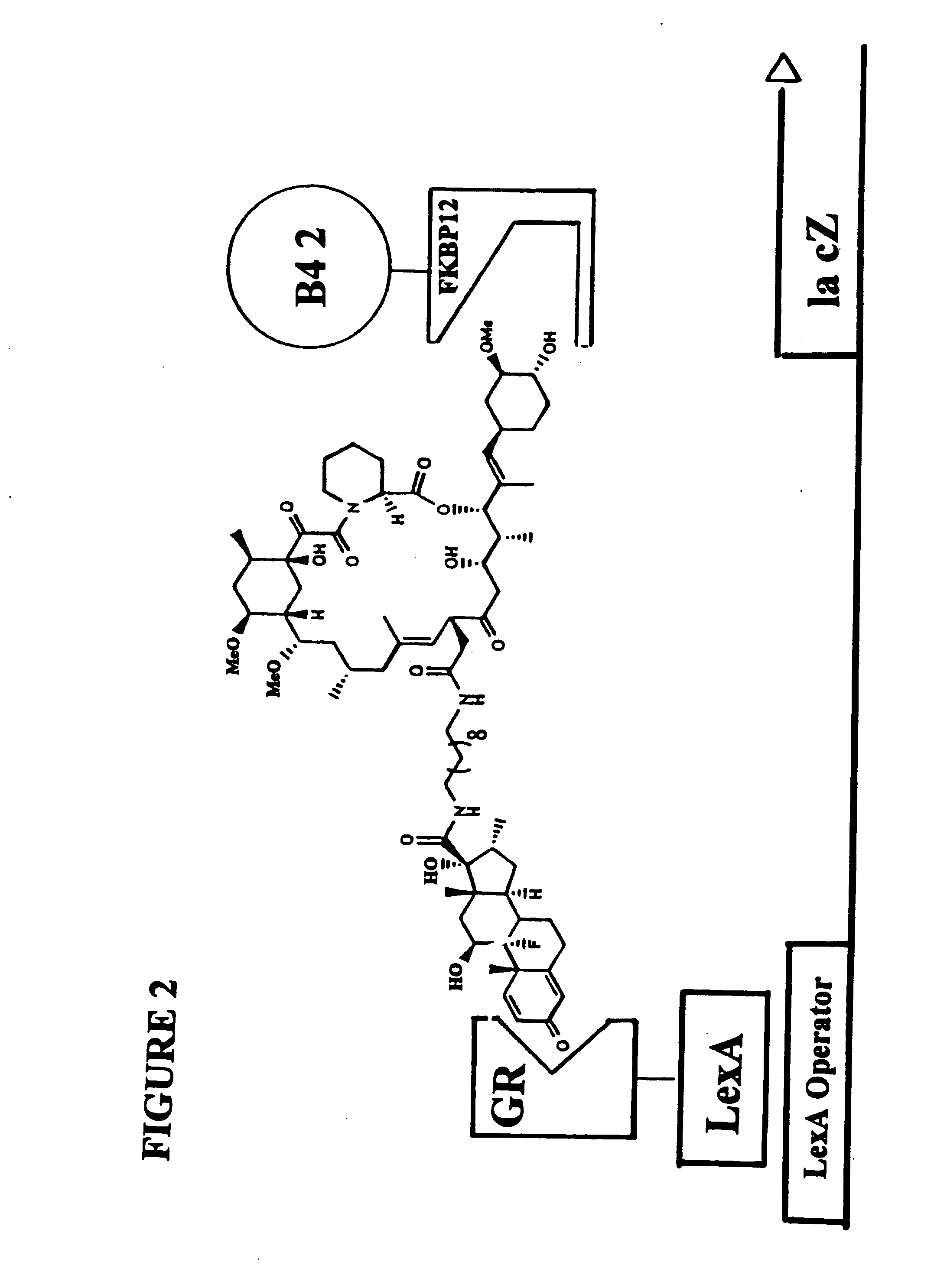
[0186]The subject invention is exemplified using the components of the yeast three-hybrid system (Licitra, represented in FIG. 2, see also U.S. Pat. No. 5,928,868). In this system DEX-FK506 (exemplifying H1-H2) mediates dimerization of the protein fusions LexA-GR (representing reporter V—H1 receptor) and B42-FKBP12 (representing reporter W—H2 receptor) thus activating transcription of a lacZ reporter gene. The chemical handles H1 and H2 and the protein dimerization assay, however, all can be varied.
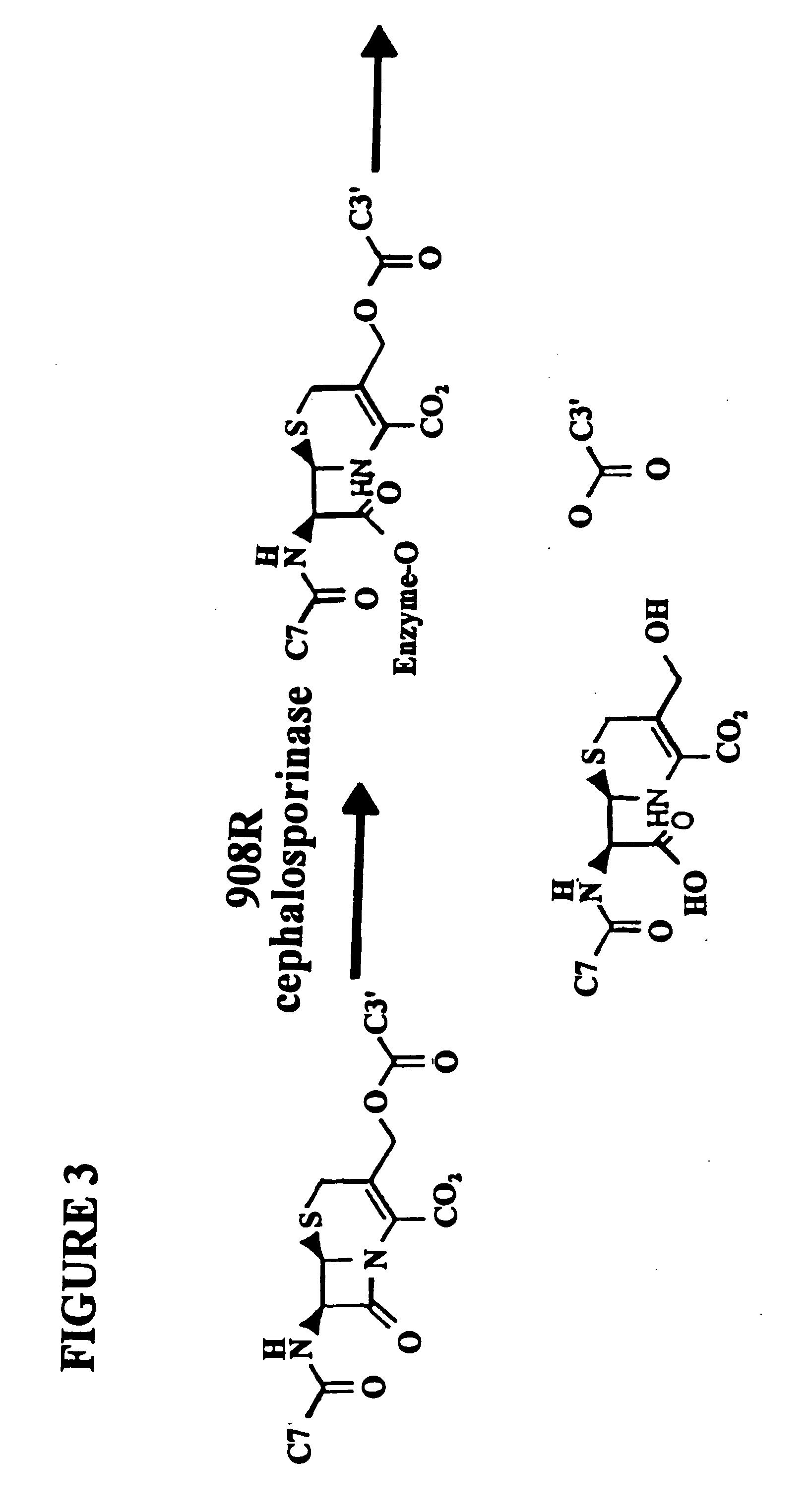
[0187]In the subject invention, however, the yeast three-hybrid system is altered by inserting a BOND, B, as well as any required spacers X and Y, so as to form a small molecule having the structure H1-X—B—Y—H2. While there is ample precedent for small-molecule mediated protein dimerization, what remains is to show these assays can be used to select for catalysts. Cephalosporin hydrolysis by a cephalospo...
example 3
[0202]CIDs can be used to screen cDNA libraries based on biochemical function. This glycosidase example is used to determine the best method for expressing the cDNA clones and to optimize the screening process.
Proof of Principle—β-Galactosidase Activity Assays
[0203]Table III explains the components of each strain. Each strain was constructed from the parent yeast strain FY250 and also contains the pMW106 plasmid, which has the LacZ reporter gene that is turned on only in when the LexA DNA binding domain and the B42 activation are brought in to the vicinity of each other. We use several different strains because we use DHFR from two different species, mDHFR is from murine, while eDHFR is from E. coli. We are asl oable to switch the small moleculebinding domains. For example, the strain containing LexA-eDHFR with B42-rGH2 is a different strain and behaves differently from the strain containing LexA-rGR2 with B42-eDHFR. We also put in short 6 amino acid linkers between the two domains ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| structure-activity relationships | aaaaa | aaaaa |
| acid and base sensitive | aaaaa | aaaaa |
| affinity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com