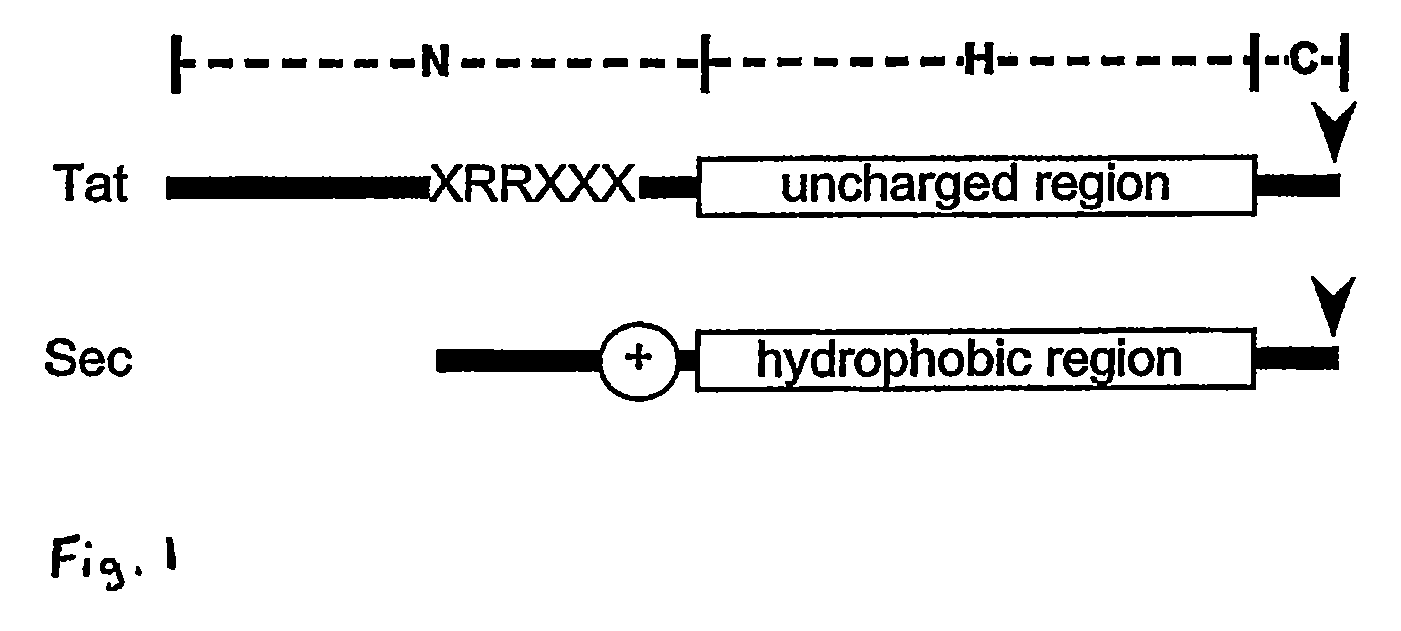
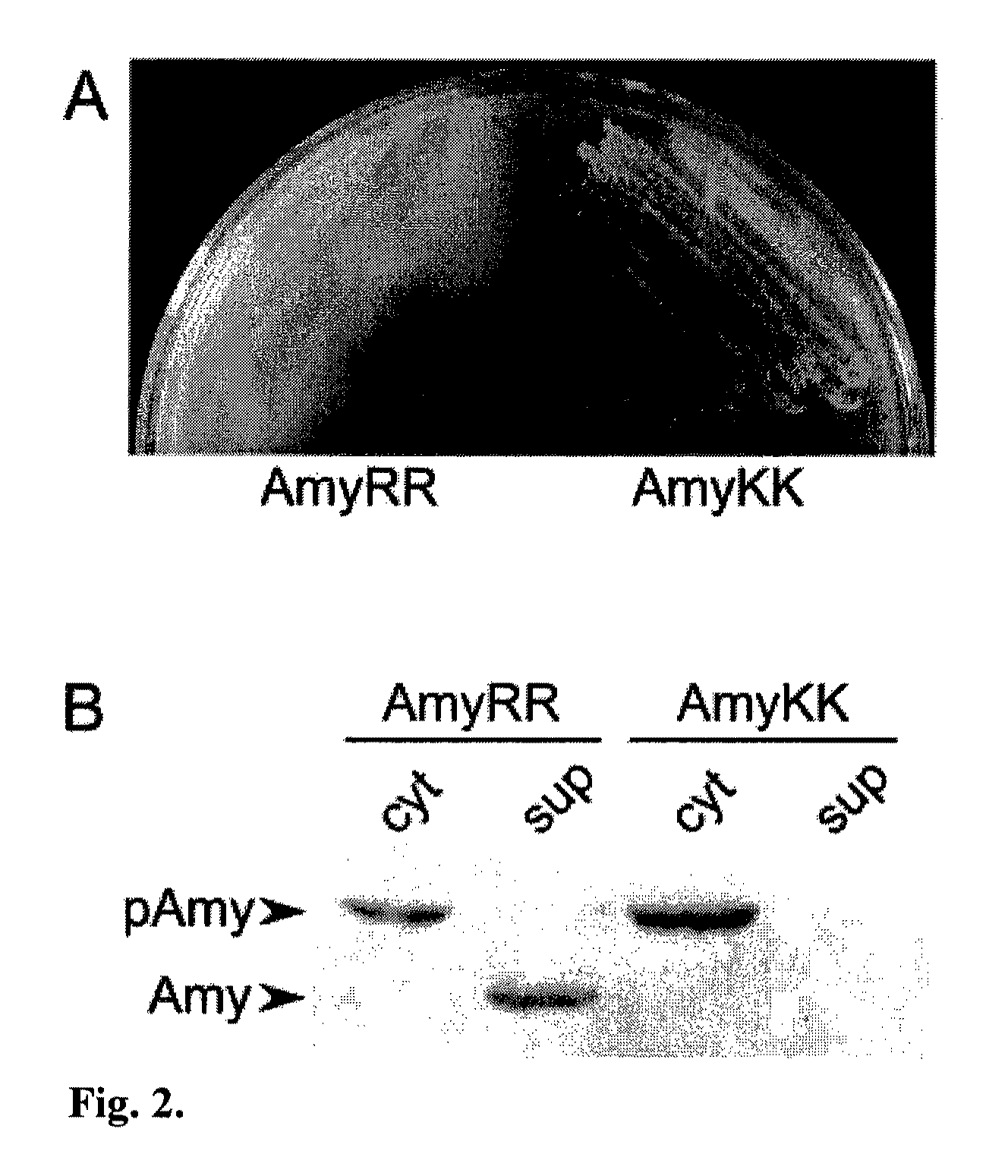
Heterologous Protein Production Using The Twin Arginine Translocation Pathway
a technology of arginine translocation and heterologous protein, which is applied in the field of heterologous protein production using the twin arginine translocation pathway, can solve the problems of difficult folding in the external environment, inability to secrete heterologous protein, and high cost of protein separation, so as to achieve the effect of purification and production
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
TATFIND 1.1
[0087]To examine utilization of the Tat and Sec pathways in Halobacteriaceae, analyses were conducted in Halobacterium sp. NRC-1, currently the only known complete genomic sequence of a halophilic archaeon. The entire genome was analyzed using the following strategy: (i) a Tat substrate recognition program (TATFIND) was developed to detect putative Tat substrates in Halobacterium sp. NRC-1 (this program, described in greater detail below, is based on the position and sequence of the Tat pattern, as well as on the position, length and hydrophobicity of an uncharged region following the twin-arginine pattern); (ii) usage of the Sec pathway in Halobacterium sp. NRC-1 was examined by identifying putative secreted proteins with SIGNALP (Nielsen et al., 1997); (iii) all SIGNALP-positive candidates were analyzed further with the TMHMM program (Sonnhammer et al., Proc. Int. Conf. Intell Syst. Mol Biol. 6:175-182 (1998)) that predicts membrane spanning segments, thereby eliminatin...
example 2
TATFIND 1.2 and TATFIND 1.3
[0113]A refined TATFIND program was used to look at all prokaryotic genomes available at NCBI. The recent identification of novel Tat substrates in P. aeruginosa (Ochsner et al., 2002) permitted the extension of the rules of TATFIND 1.1 to allow a methionine at position X−1, and glutamine at position X+4, thus creating a second program, TATFIND 1.2. Mutational analyses of certain Tat signal sequences suggested that in specific instances the substitution of lysine, asparagine, and glutamine for one of the two conserved arginines does not prevent Tat-dependent export. However, only two naturally occurring Tat substrates are known to deviate from the conserved ‘RR’ motif in their signal sequence. Therefore, modifications of the program allowing for a variable RR motif due to these recent reports were not included in the present study, as these were likely to be exceptions that would lead to strong overprediction.
[0114]Tat Substrates—TATFIND 1.2 and TATFIND 1....
example 3
Cloning of Streptomyces Tat Components and Putative Tat Substrates
[0126]To determine the role and efficiency of a Streptomyces, the preferred organism was S. lividans because as an expression host include 1) it does not secrete a blue pigment like S. coelicolor, and 2) it does not require non-methylated DNA. However, the genome for S. lividans has not yet been completely sequenced. Therefore, S. coelicolor, was selected for the following analysis because it is highly homologous to S. lividans, its genome has been fully sequenced, permitting a TATFIND analysis of the genome, and complete characterization of the genomic information for the strain was advantageous for the analysis. It is intended that the findings in S. coelicolor are also applicable to the closely related S. lividans, as well as to other Streptomyces strains.
[0127]Accordingly in vivo analyses were initiated of the Tat pathway in S. coelicolor. A His-tagged version of the S. coelicolor Tat C components were cloned into...
PUM
| Property | Measurement | Unit |
|---|---|---|
| concentration | aaaaa | aaaaa |
| degree of tertiary structure | aaaaa | aaaaa |
| energy | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com