MGAL: A GAL Gene Switch-Based Suite of Methods for Protein Analyses and Protein Expression in Multicellular Organisms and Cells Therefrom
a multicellular organism and gene switch technology, applied in the field of gene switch-based protein analysis suite of multicellular organisms and cells, can solve the problems of two-hybrid methods in multicellular organisms or cells, existing methods suffer from serious limitations, and few existing methods for detecting and analyzing protein-protein interactions, etc., to achieve increased expression, increase or decrease growth or survival, and increase expression
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Preparation of Plasmids Containing Membrane Targeting Sequences and Localization of Fusion Protein Expression in Yeast
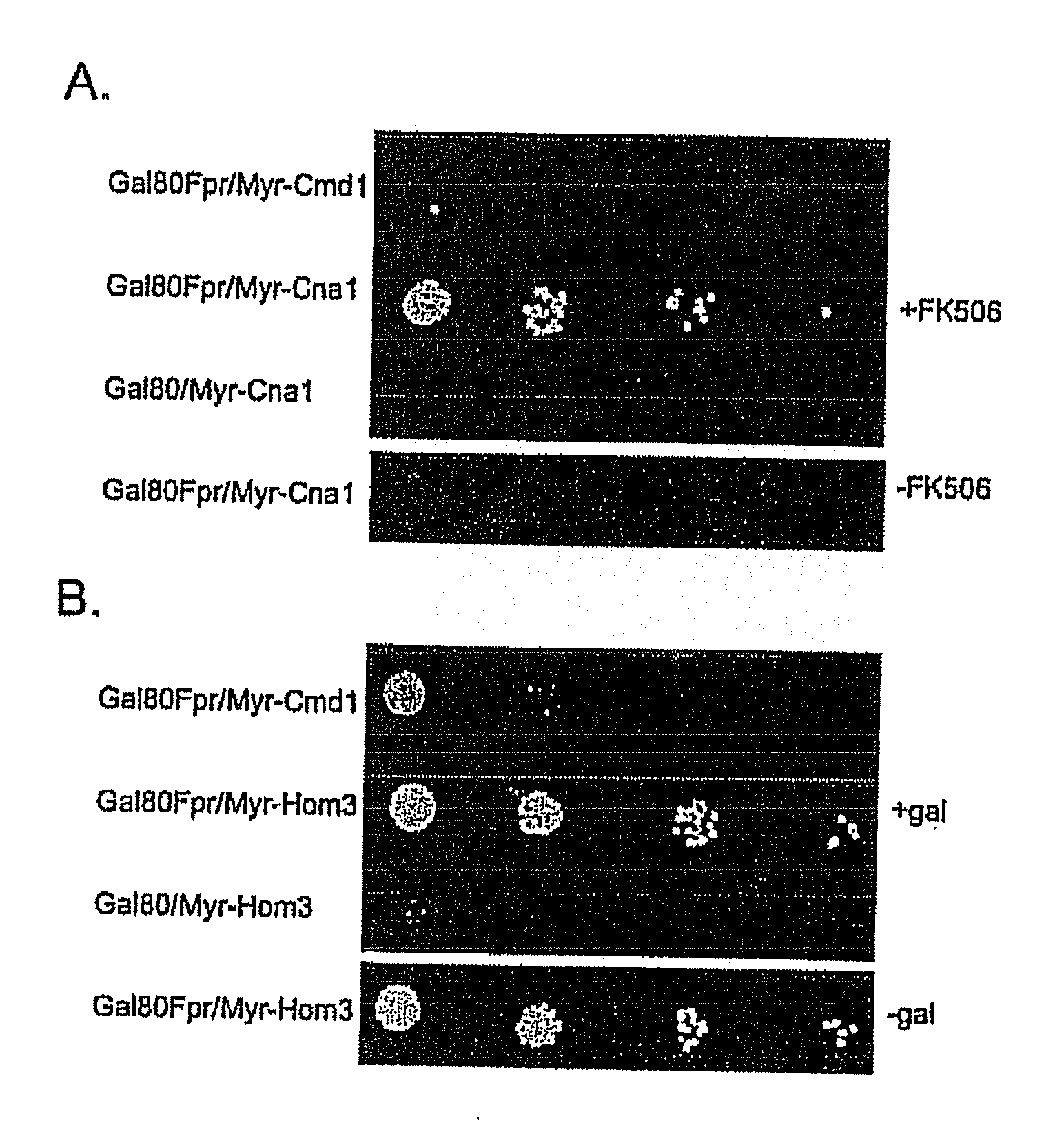
[0104]To determine if Gal80p would interact with a membrane-bound binding partner, Gal3p (Gal80p's natural binding partner) was tethered to the cell and mitochondrial membranes. In addition, the effects of Gal3p sequestration on galactose induction of GAL gene expression were examined.
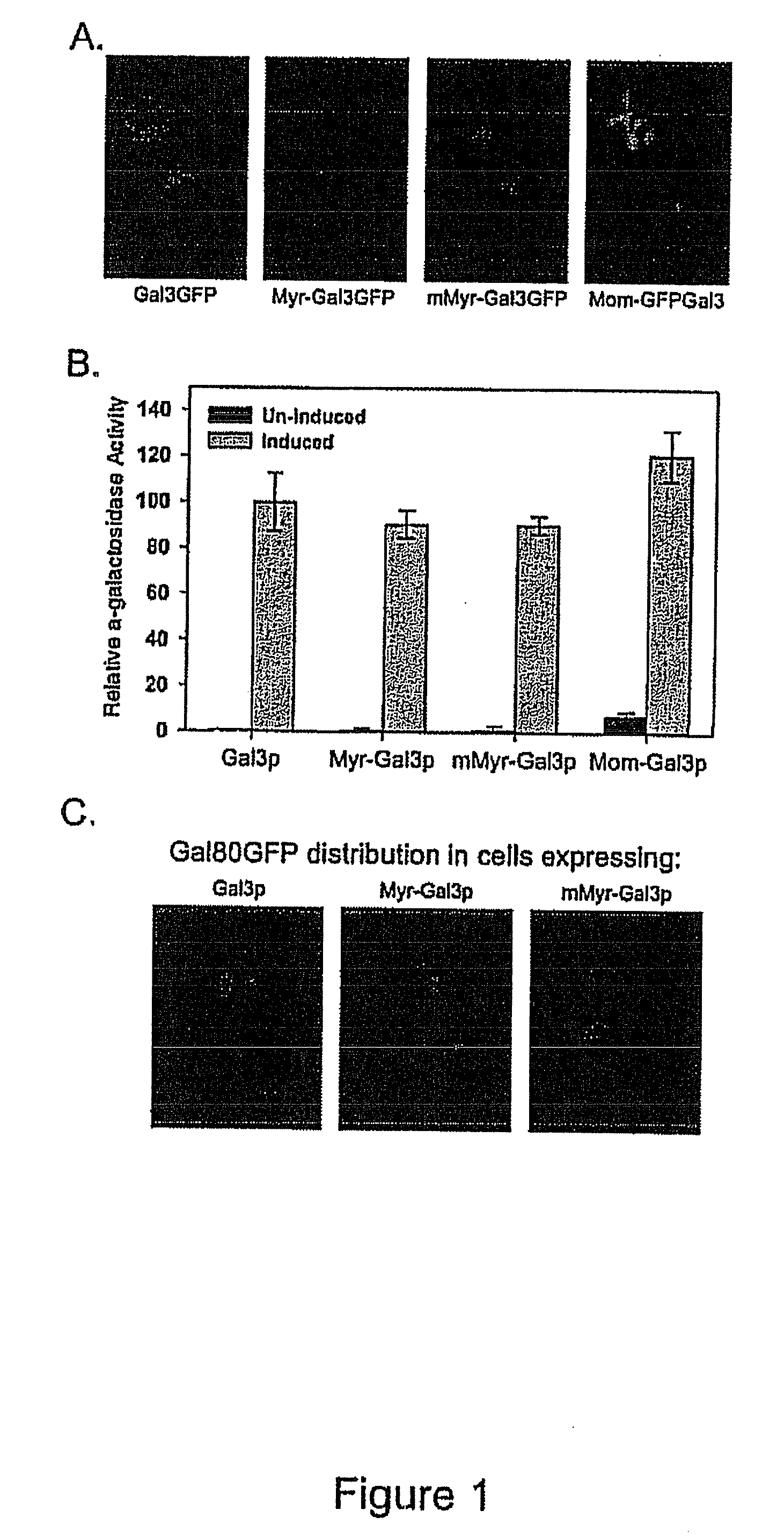
[0105]Gal3p-GFP was targeted to the cytoplasmic or mitochondrial outer membranes by fusion to a protein myristoylation signal (Myr-Gal3p-GFP) or mitochondrial outer membrane signal anchor sequence (MOM-Gal3p-GFP), respectively. N-myristoylation (Johnson et al., 1994, Annu. Rev. Biochem. 63: 869-914; Resh et al., 1999, Biochem. Biophys. Acta. 1451: 1-16) was chosen because it is a co-translational process that occurs when the nascent peptide is still attached to the ribosome. Mitochondria targeting was chosen because it appears to be a very fast and efficient process (Fujiki and Verner, ...
example 2
Membrane Associated Gal3p Retains Ability to Bind Gal80p and Induce GAL Gene Expression in Yeast
[0111]To verify appropriate activity of Myr-Gal3p or MOM-Gal3p in yeast cells harboring the plasmids encoding these fusion proteins, semi-quantitative colony growth assays and galactose-responsive reporter gene analyses were performed. Following verification of appropriate Gal3p expression, the distribution of Gal80p in Myr-Gal3p or MOM-Gal3p expressing cells was examined by fluorescence microscopy and Western blot analysis.
[0112]Gene expression was determined for two different types of GAL gene promoters in cells carrying the membrane bound and cytoplasm sequestered Gal3p. First, a sensitive and semi-quantitative colony growth assay was used to assess expression of a HIS3 reporter gene whose promoter bears four UASGAL sites. Cells of a gal3Δgal1Δ strain (Sc781) carrying Myr-Gal3p or MOM-Gal3p grew indistinguishably from cells harboring wild type Gal3p on synthetic medium lacking histidin...
example 3
Gal80p Association with Gal4p within Gene Promoters is Reduced in Yeast
[0116]Because the association of Gal80p with Gal4p at a GAL gene promoter is essential for inhibition of Gal4p in the absence of galactose, the effect of galactose (which triggers the interaction between Gal3p and Gal80p) on the amount of Gal80p complexed with DNA-bound Gal4p was determined (Torchia et al., 1984, Mol. Cell. Biol. 4: 1521-1527; Lohr et al., 1987, J. Biol. Chem. 262: 15589-15597). To determine the extent of Gal4p and Gal80p association with the UASGAL region of the GAL1 / GAL10 gene promoter, formaldehyde-based in vivo cross-linking assays were performed followed by chromatin immunoprecipitation.
[0117]Chromatin immunoprecipitations were performed as described by Braunstein et al., 1993, Genes Dev. 7: 592-604 and Kuras et al. 1999, Nature 399: 609-613. Briefly, wild type Sc723 cells (Blank et al., 1997, Mol. Cell. Biol. 17: 2566-2575) were grown to early exponential growth phase (as measured by absorb...
PUM
| Property | Measurement | Unit |
|---|---|---|
| volume | aaaaa | aaaaa |
| volume | aaaaa | aaaaa |
| v/v | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com