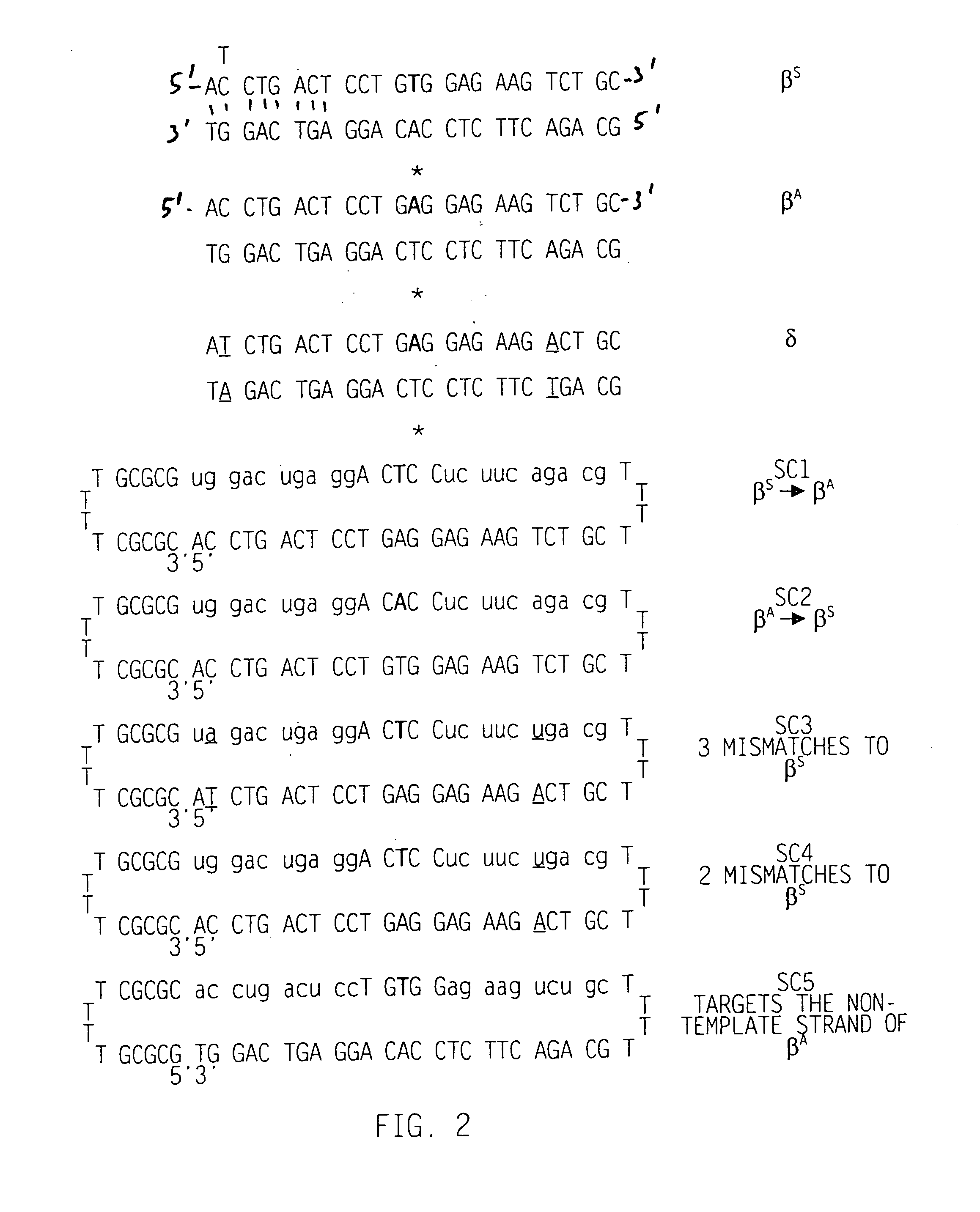Locked nucleic acid containing heteropolymers and related methods
a nucleic acid and heteropolymer technology, applied in the field of nucleic acid chimeras and methods of making single base changes, can solve the problems of limited success of conventional homologous recombination in animal models, limited success of triple-helix forming oligonucleotides coupled to cross-linking agents, and little known about the use of single-point gene modification or the introduction of cmvs in plant systems. , to achieve the effect of increasing the hybrid
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
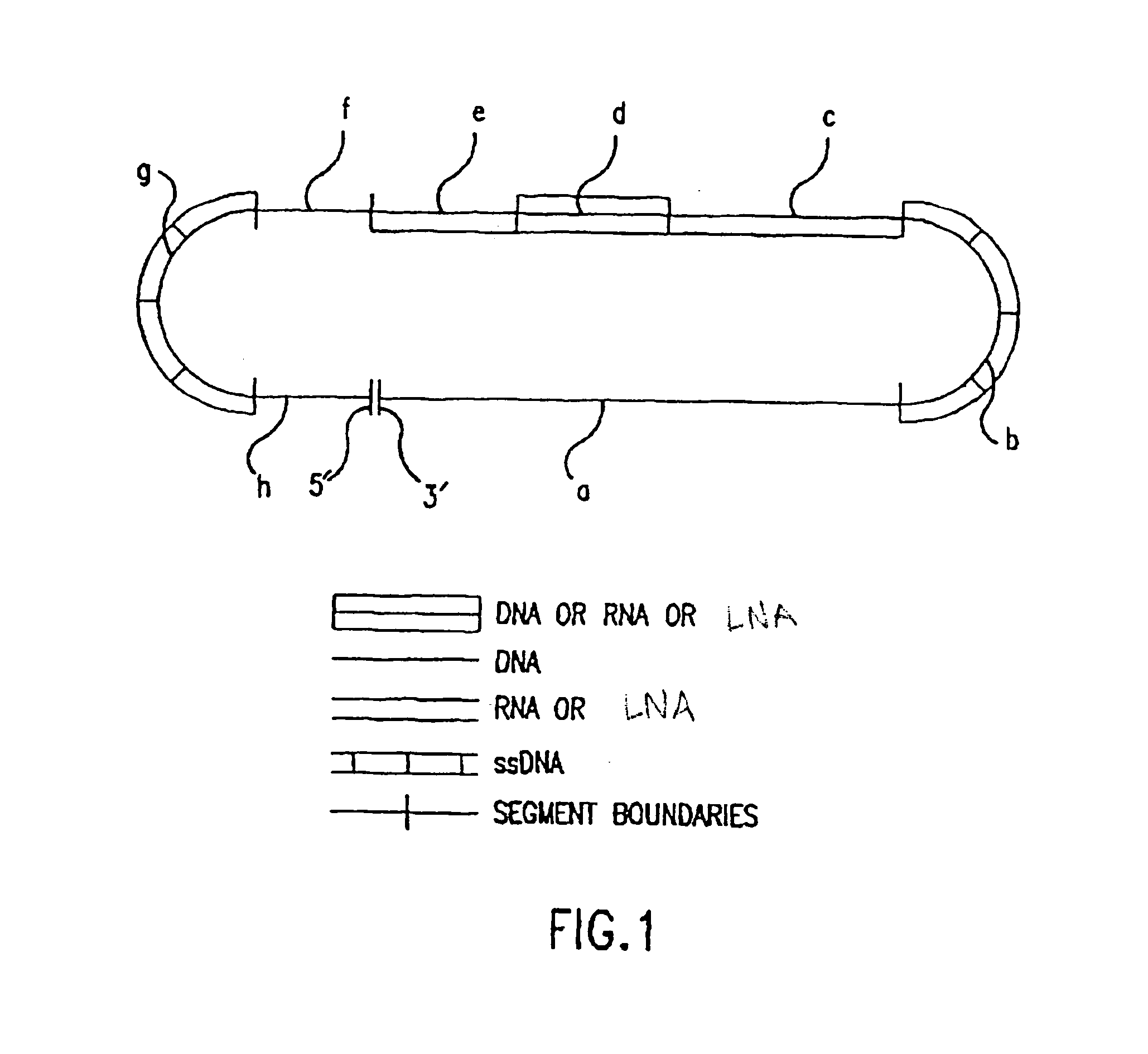
[0037] The present invention provides for compounds, termed DNA-LNA heteropolymers, that can be used to make specific changes in the genome of a eukaryotic cell. The DNA-LNA heteropolymers of the present invention may be in the form of any duplex of nucleic acids such as duplex nucleic acids, chimeric nucleic acids, heteroduplex nucleic acids, hairpin duplex nucleic acids, and / or homo-duplex nucleic acids. For definitions, see U.S. Pat. Nos. 5,565,350, and 5,792,972 incorporated herein by reference in their entirety, including any drawings.
[0038] The DNA-LNA heteropolymers of the present invention include a DNA strand and a LNA strand and optionally may include a linker. The sequence of the DNA strand and the LNA strand consists of at least two regions that are homologous to the target gene and one or more regions (“mutator regions”) that differ from the target gene and introduce the genetic change into the target gene. The mutator region is directly adjacent to homologous regions ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| lengths | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
| nucleic acid sequence | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com