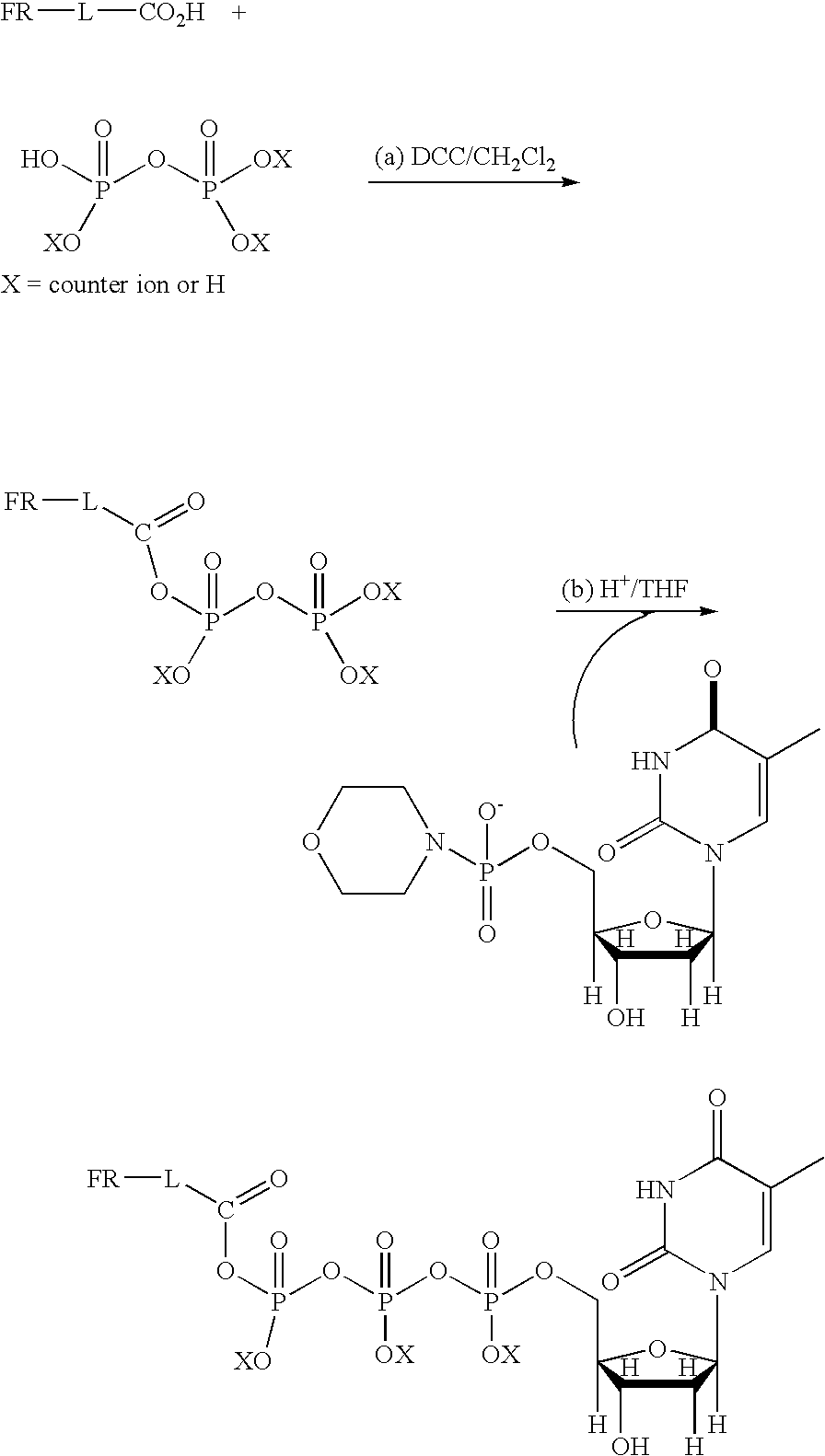
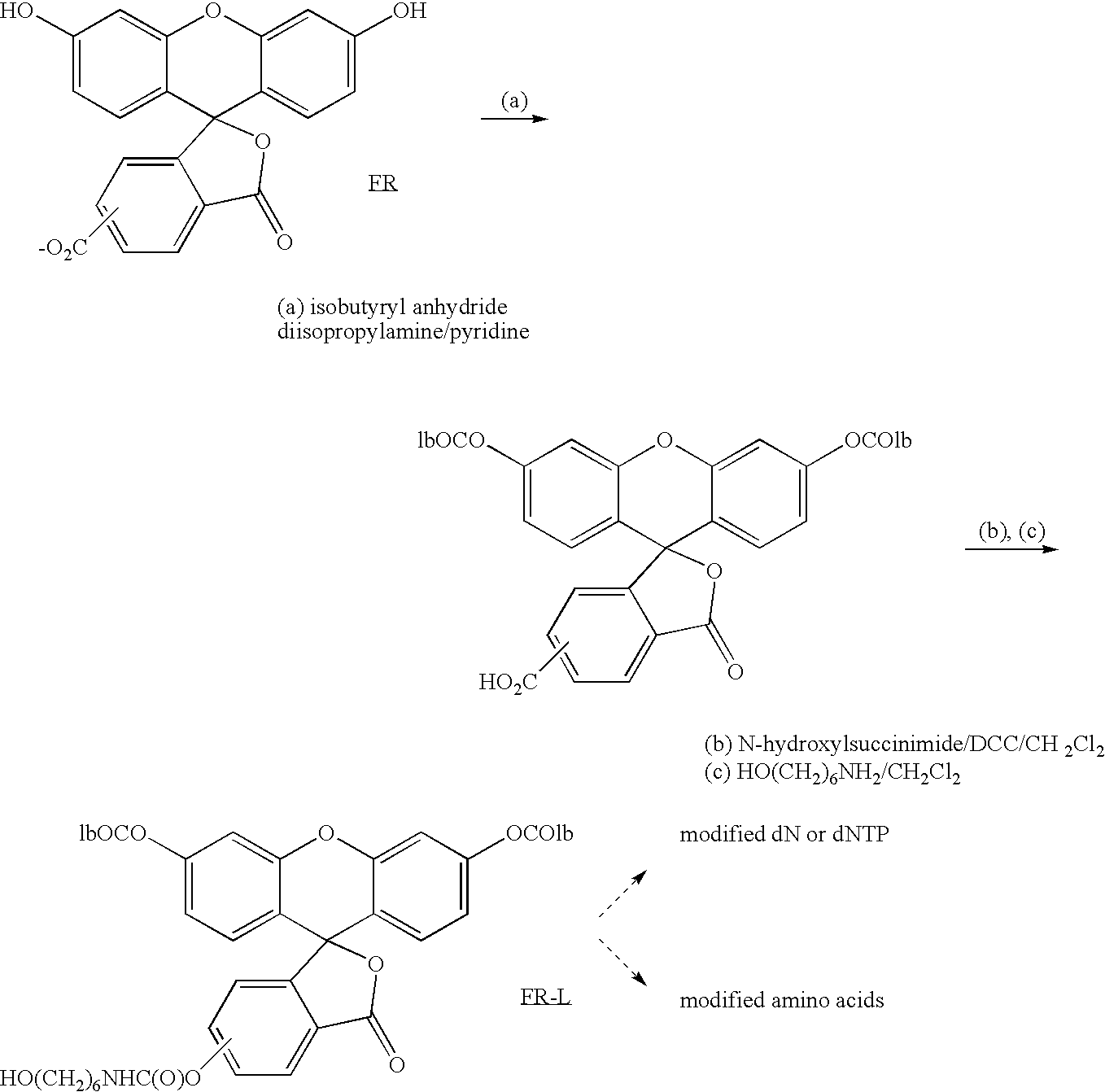
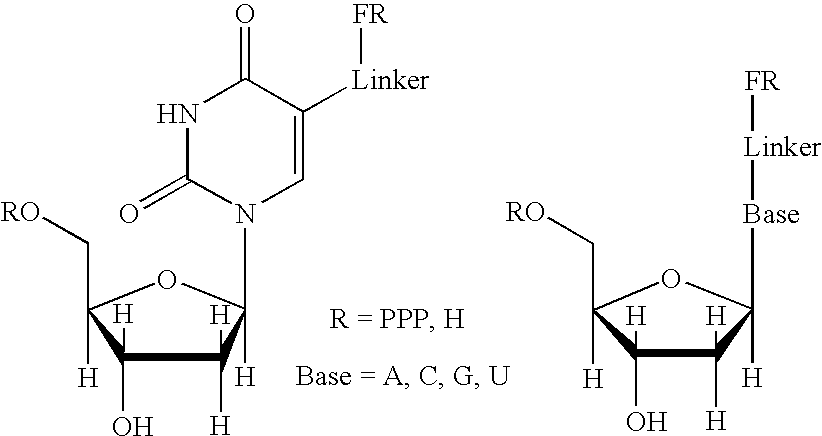
Real-time sequence determination
a single-molecule, sequence-based technology, applied in the direction of fluorescence/phosphorescence, enzymology, biomass after-treatment, etc., can solve the problem that the polymerase cannot add any additional bases to the end of the strand, the routine application of primer-walking for sequencing large dna fragments is limited, and the random approach is typically not sufficient to complete sequence determination. problems, to achieve the effect of enhancing interaction and enhancing interaction
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
examples
Cloning and Mutagenesis of Tag Polymerase
[0245] Cloning Bacteriophage lambda host strain Charon 35 harboring the full-length of the Thermus aquaticus gene encoding DNA polymerase I (Taq pol I) was obtained from the American Type Culture Collection (ATCC; Manassas, Va.). Taq pol I was amplified directly from the lysate of the infected E. coli host using the following DNA oligonucleotide primers:
Taq Pol I forward5′-gc gaattc atgaggggga tgctgcccct ctttgagccc-3′Taq Pol I reverse5′-gc gaattc accctccttgg cggagcgc cagtcctccc-3′
[0246] The underlined segment of each synthetic DNA oligonucleotide represents engineered EcoRI restriction sites immediately preceding and following the Taq pol I gene. PCR amplification using the reverse primer described above and the following forward primer created an additional construct with an N-terminal deletion of the gene:
Taq Pol I_A293_trunk5′-aatccatgggccctggaggaggc cccctggcccccgc-3′
[0247] The underlined segment corresponds to an engineered NcoI res...
PUM
| Property | Measurement | Unit |
|---|---|---|
| time | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com