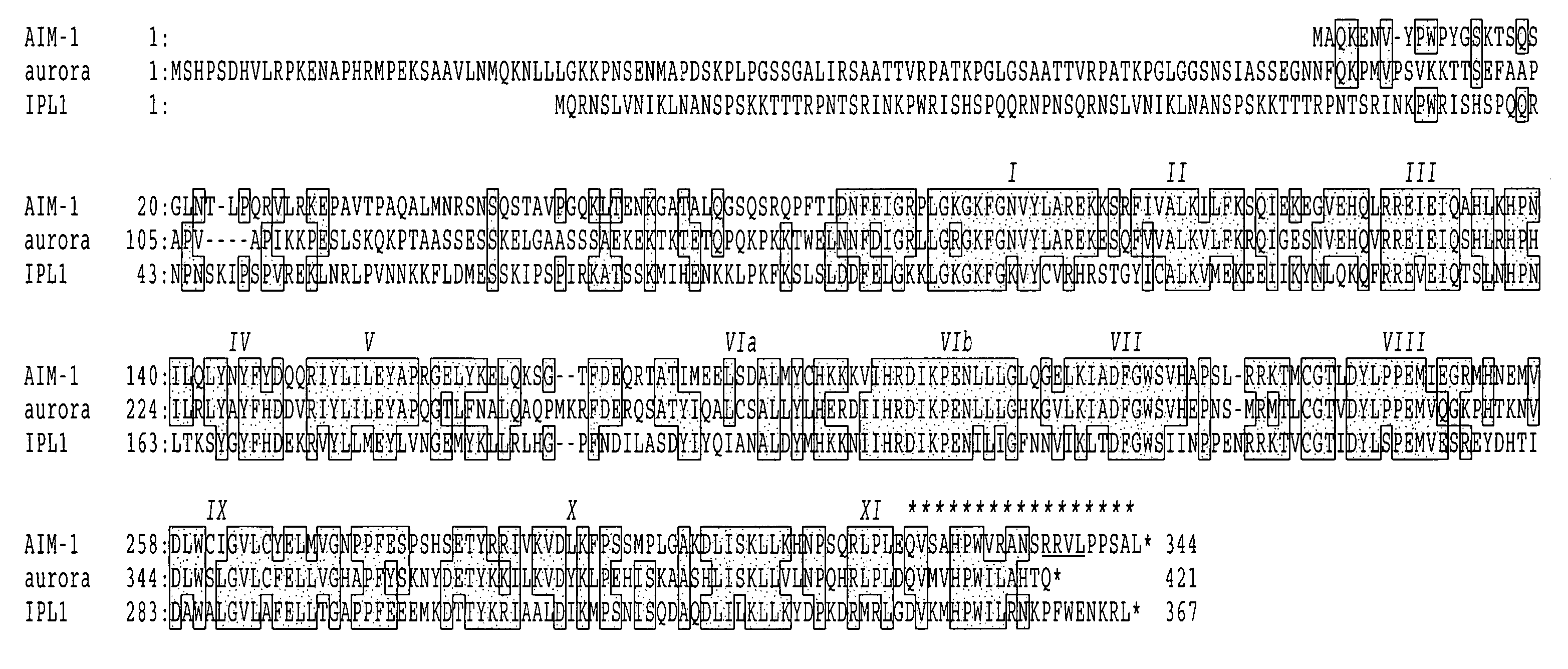
Cell cycle control protein
a cell cycle and control protein technology, applied in the direction of peptides, transferases, peptide sources, etc., can solve the problems of no molecules corresponding to aurora or ipl-1 so far in mammals, failure to adhere, cell death or oncogenesis
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
[0069] Identification of a cDNA Fragment Encoding a part of AIM-1 Gene
[0070] Oligonucleotide primer 1 sense to a conserved sequence MHRDVKP (SEQ ID NO: 3) in serine-threonine kinase domain and oligonucleotide primer 2 antisense to DFGVSGQ (SEQ ID NO: 4) were prepared. The sequences of primers 1 and 2 are as follows.
1 Primer 1: 5'- (SEQ ID NO: 5) ATGCA(T / C)(C / A)G(T / C / A / G)GA-(T / C)GT(T / C / A / G)AA(A / G)CC-3' Primer 2: 5-'- (SEQ ID NO: 6). TG(T / C / A / G)CC(T / C / A / G)GA(T / C / A / G)AC(T / C / A / G)CC(A / G)AA(A / G)TC--3'
[0071] A rat cDNA library (FEBS LETT. 320:246-250, 1993) was used as a template for amplification by 40 cycles of PCR with vent DNA polymerase using said primers 1 and 2 as primers under conditions of 94.degree. for 1 minute, 55.degree. C. for 1 minute and 720.degree. C. for 2 minutes. The PCR products were separated by agarose gel electrophoresis to give a fragment.
[0072] The CDNA fragment was sequenced to give the sequence shown as SEQ ID NO: 1 (attcacagagacataaagcccgagaacctgctgttaggtctacag...
example 2
[0073] Library Screening
[0074] (1) Preparation of a rat NRK-49F cDNA Library
[0075] NRK-49F RNA in logarithmic growth phase was extracted by guanidine method and mRNA was purified on an oligo-dT cellulose column. Oligo-dT / NotI was used as a primer to synthesize cDNA with a reverse transferase. After an EcoRI adapter was ligated, the cDNA was inserted into an expression vector pcTerraIII (FEBS LETT. 320:246-250, 1993).
[0076] (2) Screening
[0077] The sequence of the cDNA fragment obtained in Example 1 (SEQ ID NO: 1) was used as a probe for gene screening. The probe was labeled with .sup.32-P and hybridized with the NRK-49F cDNA library coupled to filters to isolate positive clones under the following conditions. Hybridization solution: 6.times.SSPE, 0.5% SDS, 10.times.Denhardt solution, 100 u / ml denatured herring sperm DNA. The filters were washed with 2.times.SSC, 0.1% SDS for 15 minutes and 0.2.times.SSC, 0.1% SDS for 15 minutes.
[0078] Thus, three full-length cDNA clones were isolated...
example 3
[0080] Expression of AIM-1 Gene in Rat Tissues
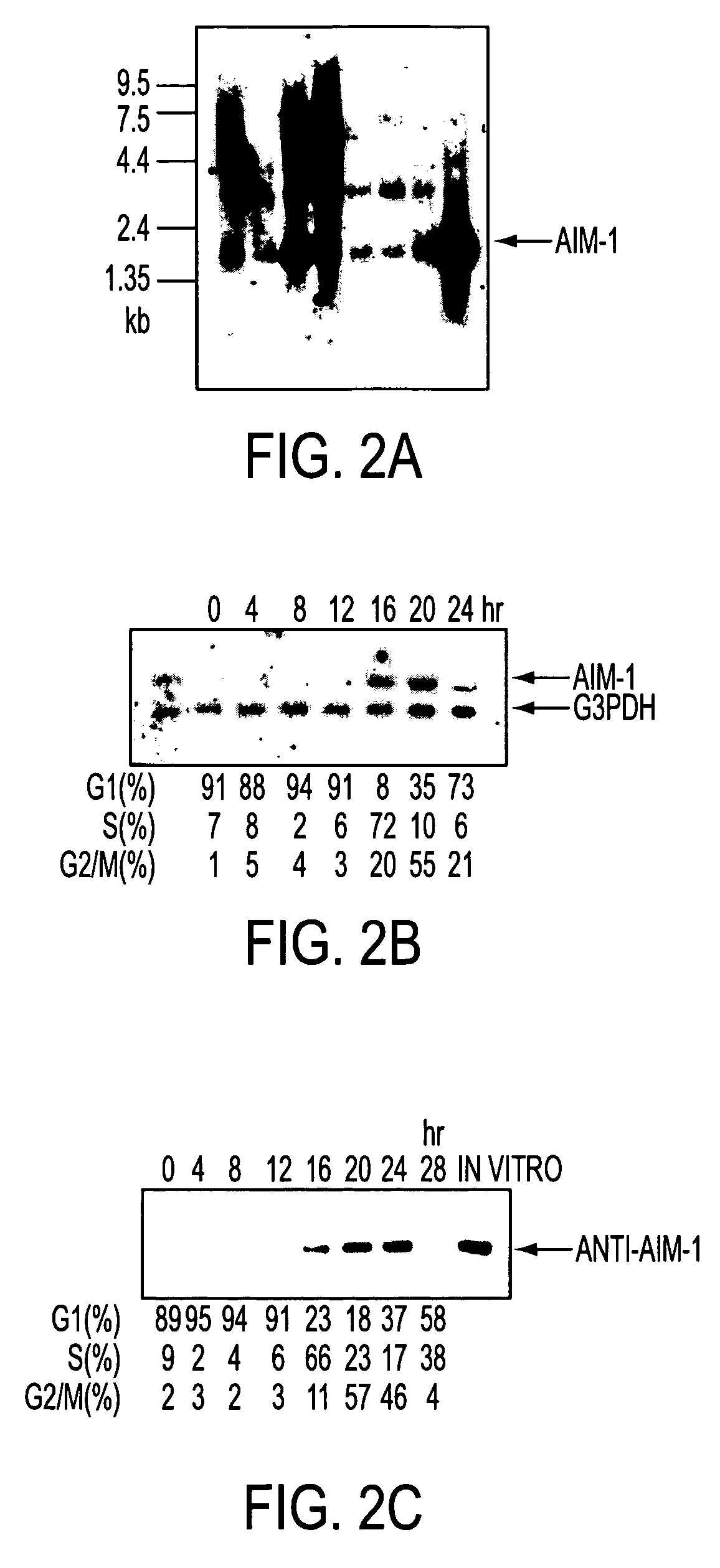
[0081] Membrane filters containing mRNA (2 .mu.g) prepared from various rat tissues were subjected to northern blot analysis with the .sup.32P-labeled AIM-1 cDNA fragment described in Example 2 as a probe.
[0082] Results are shown in FIG. 2a. A band of AIM-1 of about 2.0 kb was detected in all the tissues tested, particularly abundantly in testis, spleen and lung.
PUM
| Property | Measurement | Unit |
|---|---|---|
| pH | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
| Northern blot analysis | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com