Mutant xylose isomerase and its gene and application
A xylose isomerase and amino acid technology, which is applied in the fields of enzyme genetic engineering and enzyme biochemical engineering to achieve the effect of improving activity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
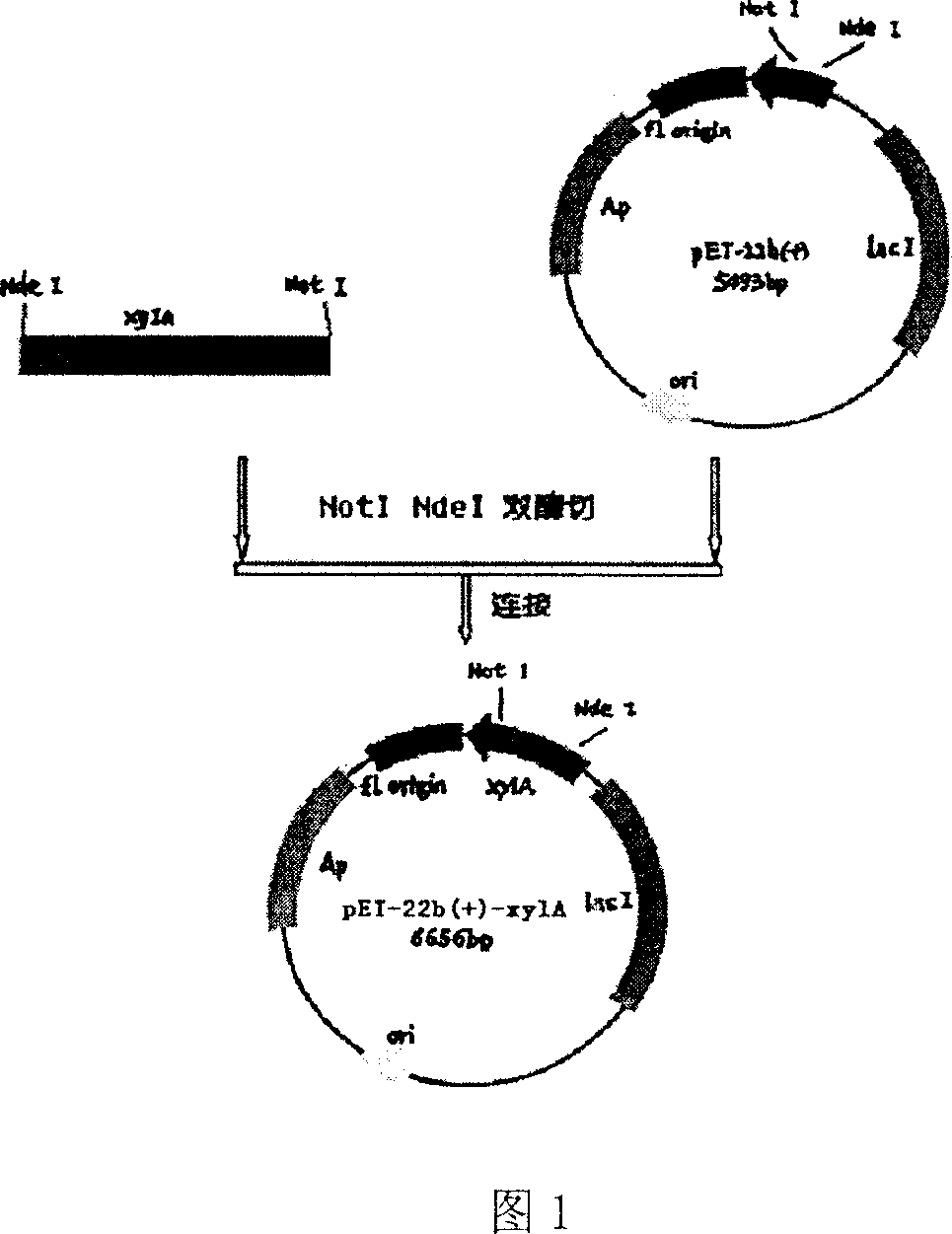
[0027] Example 1: Construction, screening and identification of recombinant plasmids expressing wild-type Thermus thermophilus HB8 xylose isomerase.
[0028] Thermus thermophilus HB8 was purchased from the American Type Culture Collection (ATCC), ATCC number 27634. Vector pET-22b(+) was purchased from Novagen.
[0029] Yeast powder 4g / L, peptone 8g / L, NaCl 2.0g / L, adjust the pH to 7.0, and autoclave at 121°C for 15min. Thermus thermophilus HB8 was inoculated in the above medium, placed on a shaker at 75°C, 200rpm, and cultured for 12-16 hours. 8000rpm, centrifuge for 15min, collect the bacteria.
[0030]The genome of Thermus Thermophilus was extracted using a genome extraction kit (Shanghai Huashun Bioengineering Co., Ltd.). Using the total DNA of Thermus Thermophilus HB8 as a template, use the following PCR primers (synthesized by Shanghai Boya Company):
[0031] Forward primer: 5'-CCATATGTACGAACCAAAACCGGAACATCGCTTTACCTTT-3' (SEQ ID NO.9)
[0032] Reverse primer: 5'-AAGG...
Embodiment 2
[0035] Example 2: Site-directed Mutagenesis of Xylose Isomerase Gene
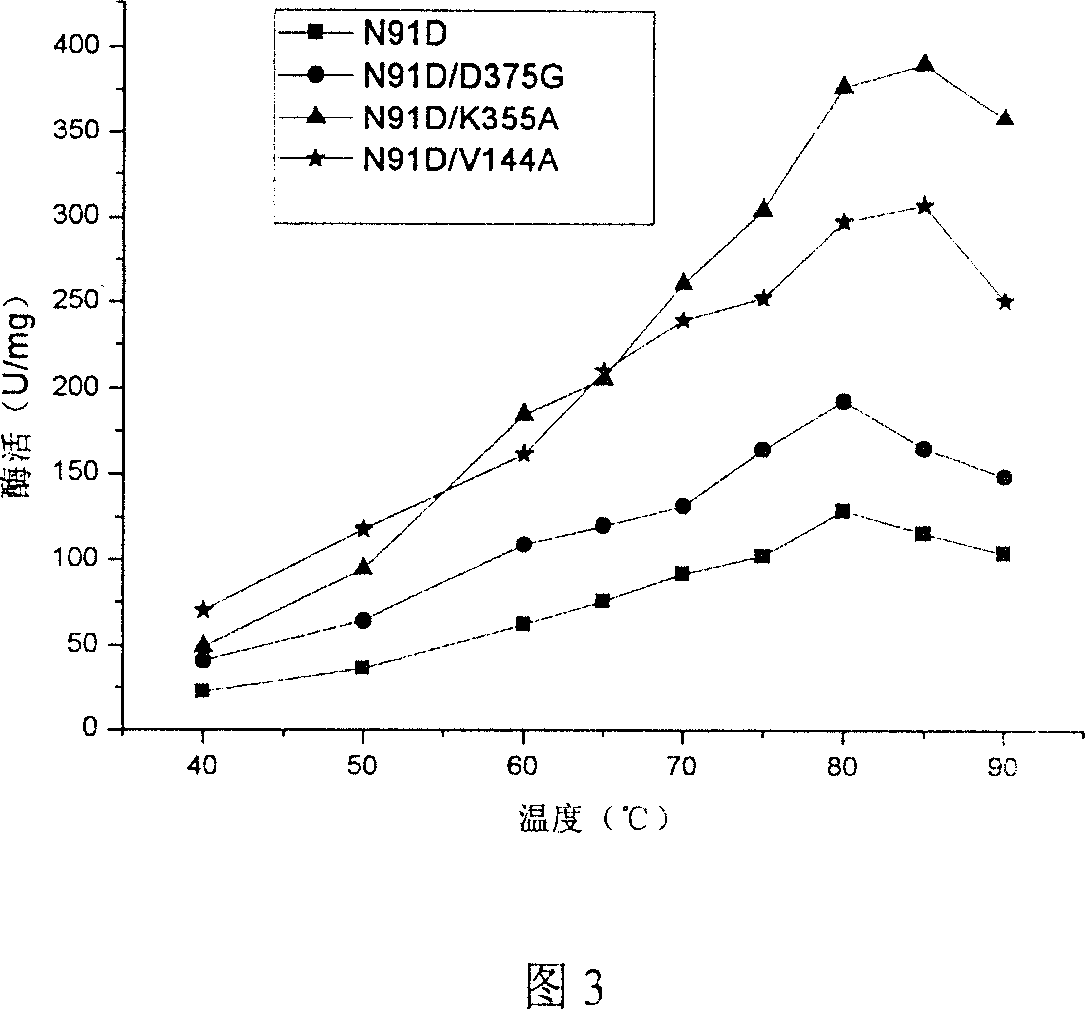
[0036] According to the requirements of the multi-point mutation kit (QuikChange® Multi Site-Directed Mutagenesis Kit, purchased from STRATAGENE Company), design the N91D primer, 5'-ATGGTCACCGCC GAC CTCTTCTCCGAC-3'(SEQ ID NO.11) was carried out by PCR using the pET-22b(+)-xylA plasmid as a template to mutate Asn91 into Asp91. According to the instructions of the multi-point mutation kit, the original plasmid template pET-22b(+)-xylA was digested with restriction endonuclease Dpn I, and the PCR product was transformed into competent cells with high efficiency. The transformed competent cells were cultured using conventional genetic engineering techniques, the plasmids were extracted for sequencing and identification, and the recombinant plasmid pET-22b(+)-xylA-N91D containing the mutated gene was obtained.
[0037] Using D375G primer 5'-TGGAACGCCTG GGC CAGCTGGCGGTG-3' (SEQ ID NO.12), PCR was carried out u...
Embodiment 3
[0040] Example 3: Expression of mutant xylose isomerase in Escherichia coli Rosseta
[0041] Transform the recombinant plasmid containing the mutant gene prepared in Example 2 into the commercialized expression host Escherichia coli Rosetta (DE3), pick a single colony into the LB culture fluid containing 75 μg / mL ampicillin and 34 μg / ml chloramphenicol , cultivate overnight at 37°C with shaking, then inoculate into LB medium containing 75 μg / mL ampicillin and 34 μg / mL chloramphenicol at 2% (v / v) inoculum size, and cultivate to OD at 37°C 600 At about 0.6, add IPTG to a final concentration of 0.9 mM, induce expression for 7 hours, take a certain volume of bacterial liquid at 8000 rpm, and centrifuge at 4°C for 15 minutes to collect the bacterial cells. The SDS-PAGE analysis of the expression product is shown in Figure 2.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com