Synthetic lethal screen using RNA interference
A technology with small interference and effect, applied in the field of lethal/co-lethal screening, cell response genes, and can solve problems such as increasing the induction of apoptosis
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
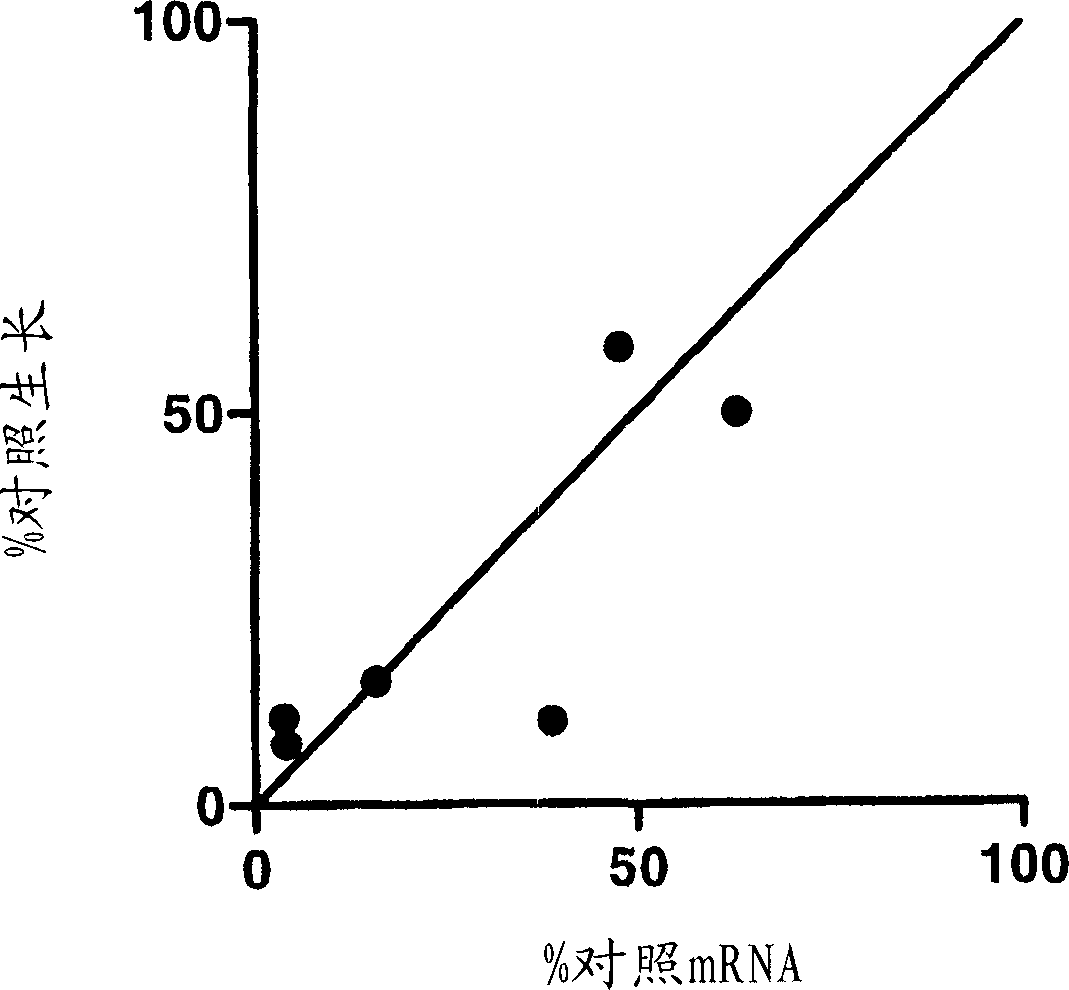
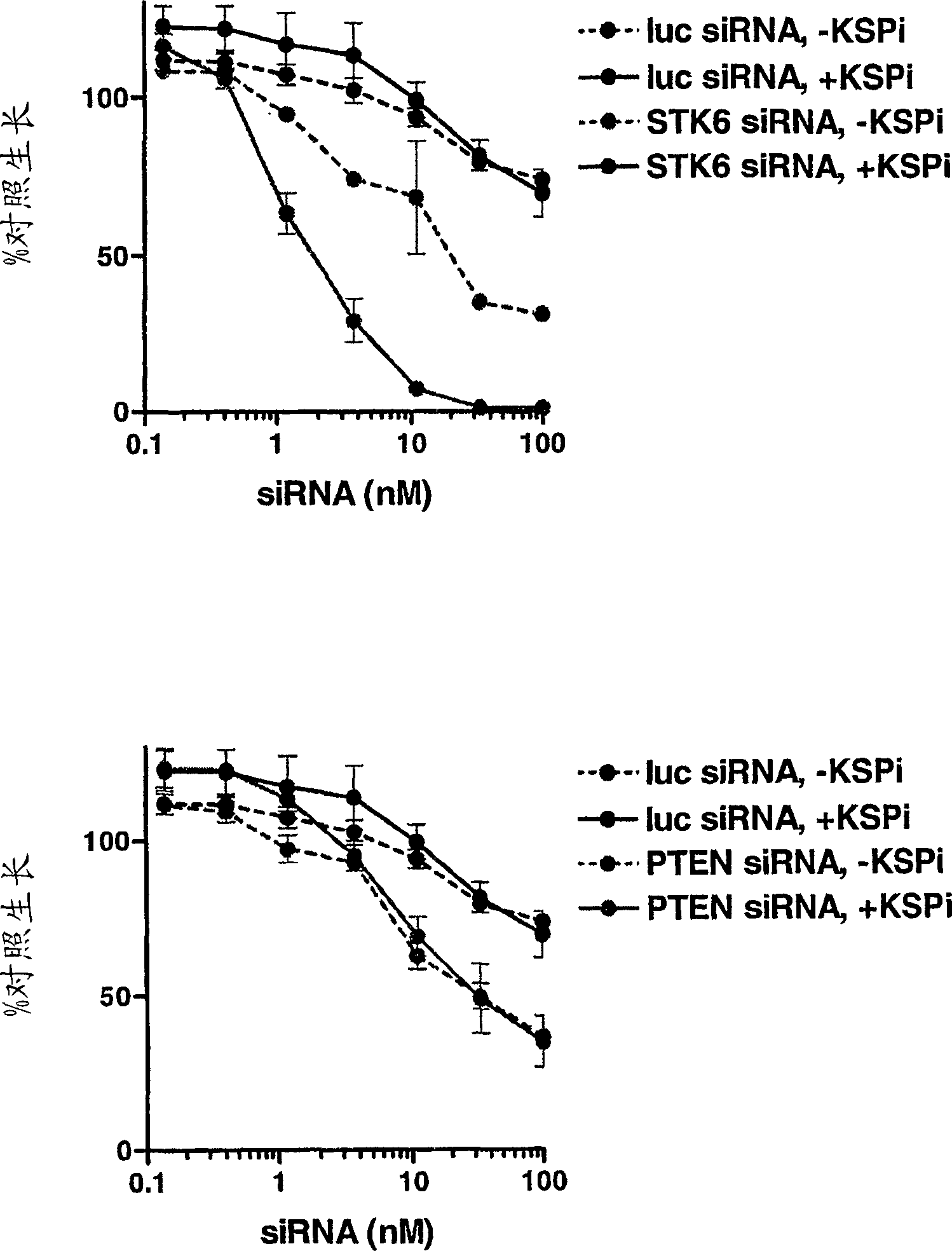
[0440]6.1. Example 1: STK6 and TPX2 and KSP interactions
[0441]This example illustrates a gene that interacts with the KSP gene inhibitor in the screening of the siRNA library. CIN8 is a beer glymeted origin of KSP. The lack of Cin8 deletion mutant is survive and has identified a must-have (Cin8 existence) (GEISER ET Al., 1997, Mol Biol Cell.8: 1035-1050) . Similarly, the destruction of human homologs in which these genes may be inferred in the presence of sub-optimal concentrations of KSPI, which is more destructive to tumor cell growth compared to their absence conditions. The screened 11 genes containing a collaborative death of CIN8: CDC20, ROCK2, TTK, FZR1, BUB1, BUB3, BUB1B, MAD1L1, MAD2L1, DNCH1, and SiRNA library of homologous siRNA libraries in siRNA libraries in siRNA libraries ( 1S) -1 - {[2S) -4- (2,5-difluorophenyl) -2-phenyl-2,5-dihydro-1H-pyrrole-1-yl] carbonyl} -2-A Cylindlamine (EC50Genes of the effect of about 80 nm. The sequence of siRNA of the 11 genes of the targ...
Embodiment 2
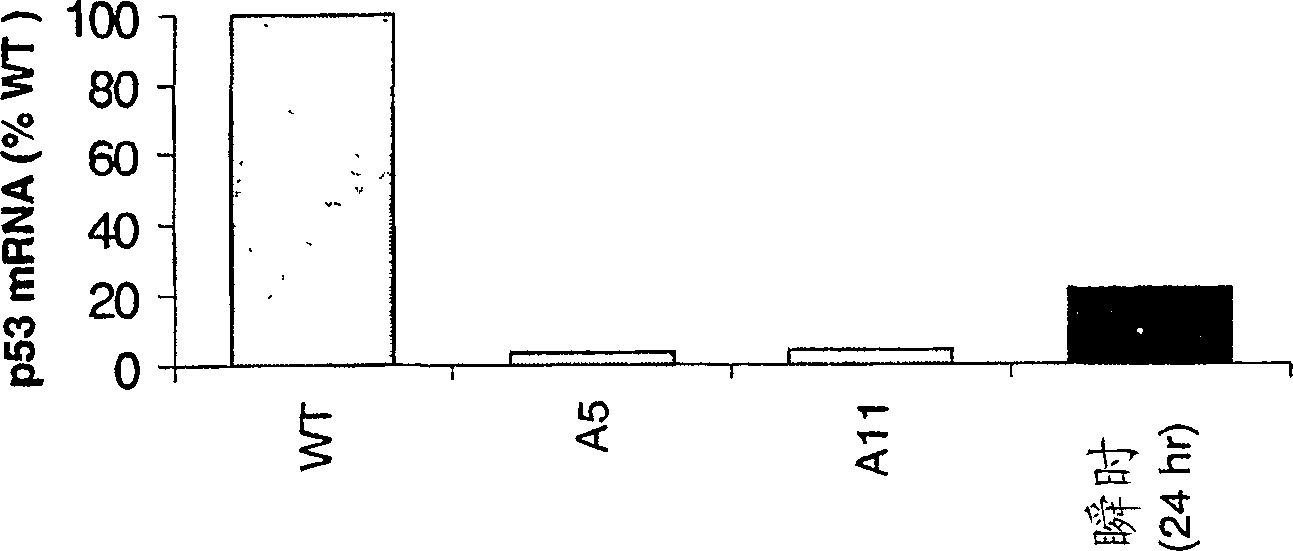
[0449]6.2 Example 2: Collaborative lethal screening with shRNA and siRNA
[0450]In this embodiment, the CHEK1 and TP53 are simultaneously mediated silencing, resulting in synergistic fatal effects in human tumor cells.
[0451]Two problems limits the potential of collaborative lethal screening with RNAi methods. First, the demonstration of collaborative death needs to be observed in a cell tend to be observed in a cell that is deleted or mutated in the first hit gene, and observed in cells containing only wild-type alleles or proteins. . Therefore, for highly controlled experiments, it is necessary to use the matching cell line of the same gene in the first hit gene damage to the measurement to solubilize. For mostly available tumor cell lines, the matching cell line cannot be obtained. Secondly, attempts to produce two gene destruction in cells in cells are hindered by the observation of the observation of the SiRNA of different mRNAs, which is used, effectively reduces the effectivenes...
Embodiment 3
[0459]6.3. Example 3: Genes enhanced or attenuated cell killing genes caused by DNA destructors
[0460]This example illustrates a semi-automatic siRNA screening for identifying a gene of a cell kill caused by reinforcing or attenuating DNA destructors. The semi-automated platform enables the filtering of Loss-flanction RNAI with small interference RNA (siRNA's). A library of siRNA of approximately 800 kinds of human genes was used to identify DNA destroyer doxorubicin (DOX), hikampto and cisplatin (CIS) enhancers. In each screening, many genes ("hits") were identified, which destroyed cellular killing of the chemotherapeutic agent. (See Table IIIA-C). Some of these hits (such as WEE1) suggest to enhance new targets for common chemotherapeutic agents; other hits (BRCA1, BRCA2) suggest new therapy for cancer caused by mutations that determine these genes.
[0461]In order to perform gene screening in human cells, a library of siRNA double strands was formed. One form of the library targets...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com