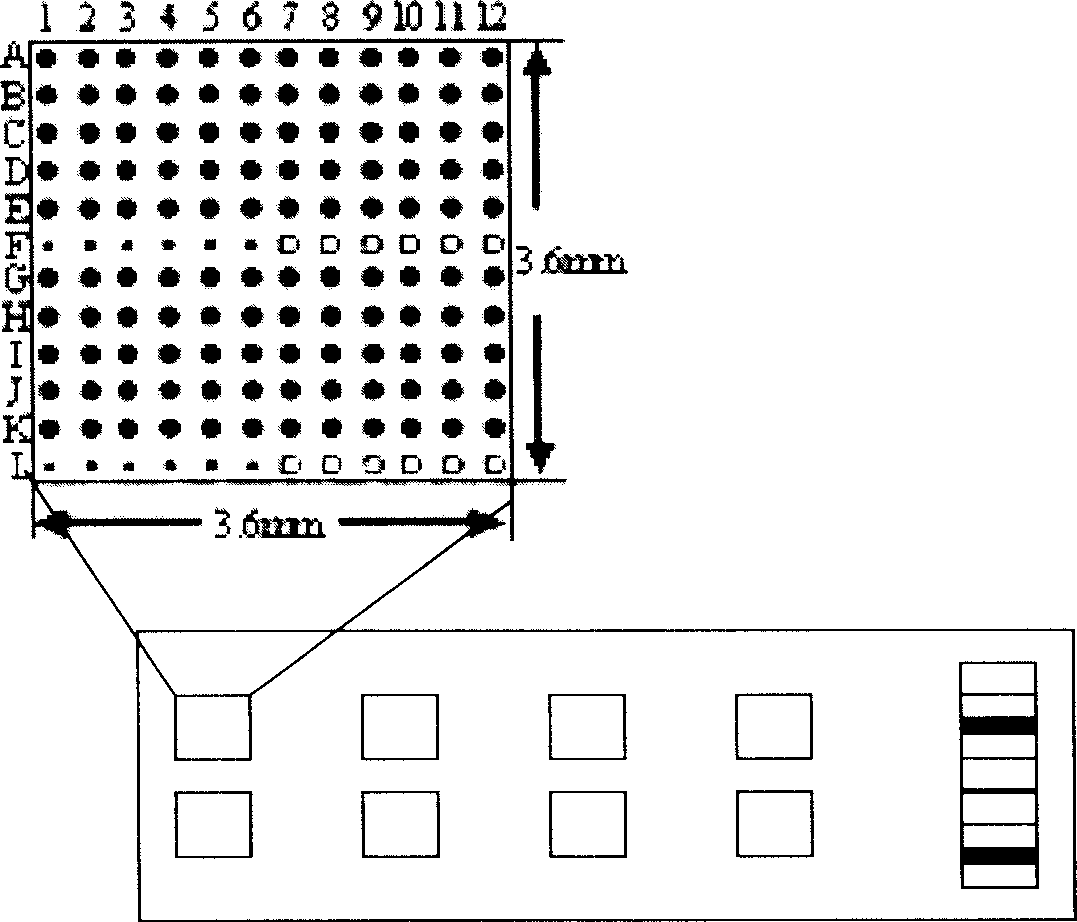
Quick detection method of pathogenic microbe diagnosis type gene chip
A technology of pathogenic microorganisms and gene chips, which is applied in the field of rapid detection of pathogenic microorganisms diagnostic gene chips, can solve the problems of lowering the annealing temperature, establishing correlation relationships, poor control of hybridization kinetics, etc., achieving improved labeling efficiency and simplified operations Stable effect of steps, hybridization effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0038] The present embodiment takes the HBV-DNA diagnostic chip as an example, and its detection method comprises the following steps:
[0039] (1) Preparation of slides:
[0040] With NaOH-ethanol (70gNaOH, 300mLddH 2 (O, 500mL 95% frozen ethanol) to clean the slides for 2h, wash with water several times, and then use poly-L-Lysine solution (70mL poly-L-Lysine, 70mL PBS for tissue culture, 600mL ddH 2 O) Coating was carried out for 1 hour, washed with water for 5 times, dried at 45° C., and placed at room temperature for two weeks for use.
[0041] Cut the coverslip into an appropriate size, wash with 2% SDS for 10min, ddH 2 Rinse with O, dehydrate with 100% ethanol, and then coat with silylating reagent; air dry for later use.
[0042] (2) Probe preparation:
[0043]Primers were designed to amplify each ORF of double-stranded HBV-DNA. There were 20 genes and full-length sequences in total. The concentration was adjusted to 0.5 μg / μL, and the same volume of DMSO was added...
Embodiment 2
[0099] In this example, HCV, HIV1G, and HPV16 subtype diagnostic gene chips and HBV, HDV, and HIV combined diagnostic chips were selected for rapid detection. The steps are carried out according to the basic steps in Example 1. The 2× labeled PCR master mix, PCR reaction system, and fast hybridization buffer are all implemented according to the formula described in the present invention. The specific operation process is basically the same as in Example 1, with the main difference It lies in designing primers to amplify probes according to different pathogen DNAs, that is, optimizing from aspects such as probe length, annealing temperature consistency, and specificity, and then taking clinical samples or purified pathogen DNA to design primers, and using the markers of the present invention Buffer for full-length labeling or specific fragment labeling. That is, different pathogens have different probes, different labeled fragments, and different PCR amplification conditions, b...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com