Nucleotide sequence encoding defence response gene of sea island cotton, plant expression vector and plant cell containing the sequence
The technology of plant expression vector and nucleotide sequence is applied to the nucleotide sequence encoding the defense response gene of sea island cotton, the plant expression vector containing the sequence and the field of plant cells, which can solve the limitation of rapid mutation of pathogenic bacteria and conventional breeding. , antigen deficiency, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0070] Embodiment 1: the preparation of the structural gene of coding GbRAR1, GbSGT1, GbRac1 protein
[0071] Total RNA was extracted from the leaves of Cotton 7124 (National Germplasm Bank of Chinese Academy of Agricultural Sciences, #00070074) treated with Verticillium dahliae, and the first strand of cDNA was synthesized with 5 μg of total RNA and oligo d(T) primer as substrate For specific operations, refer to the kit instructions (GIBCOBRL Company, catalog number #11146-024).
[0072] Cloning of 1.1GbRar1 gene
[0073] 1.1.1 Cloning of the 5' end of the GbRar1 gene
[0074] According to the cotton EST clone (GenBank accession number: BQ403789 , BF276058 , BG443488 , BG446854 , BQ412690 ) and upland cotton EST clones (GenBank accession number: AW187113 , AW187078 ), design primers:
[0075] pRR: 5'ATGGCGGAGCAAGCTGTT 3'
[0076] pFR: 5'ATTCCTCCTCATGCTTATGAC 3'
[0077] The first strand of sea island cotton cDNA was used as a template for PCR amplification....
Embodiment 2
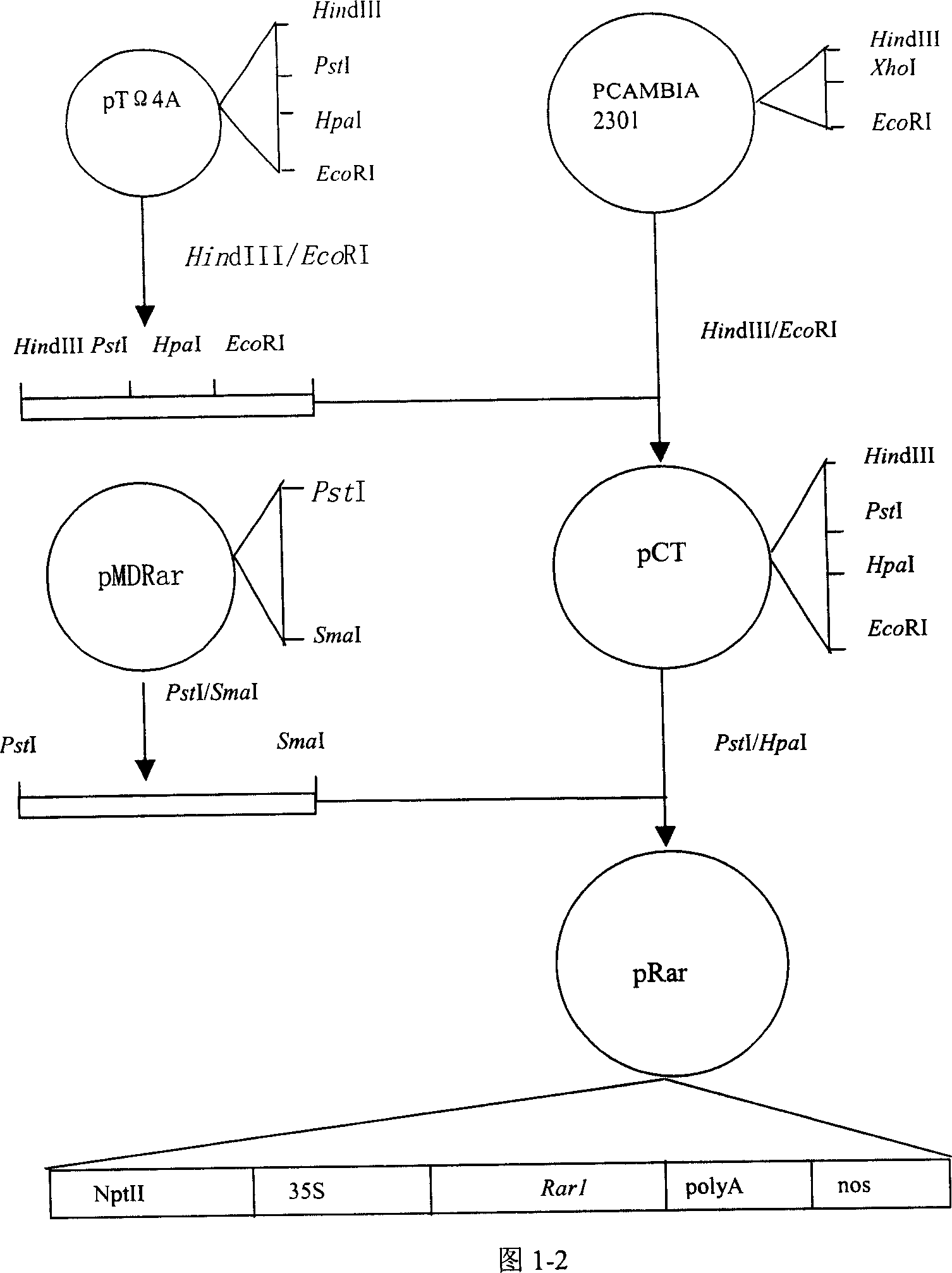
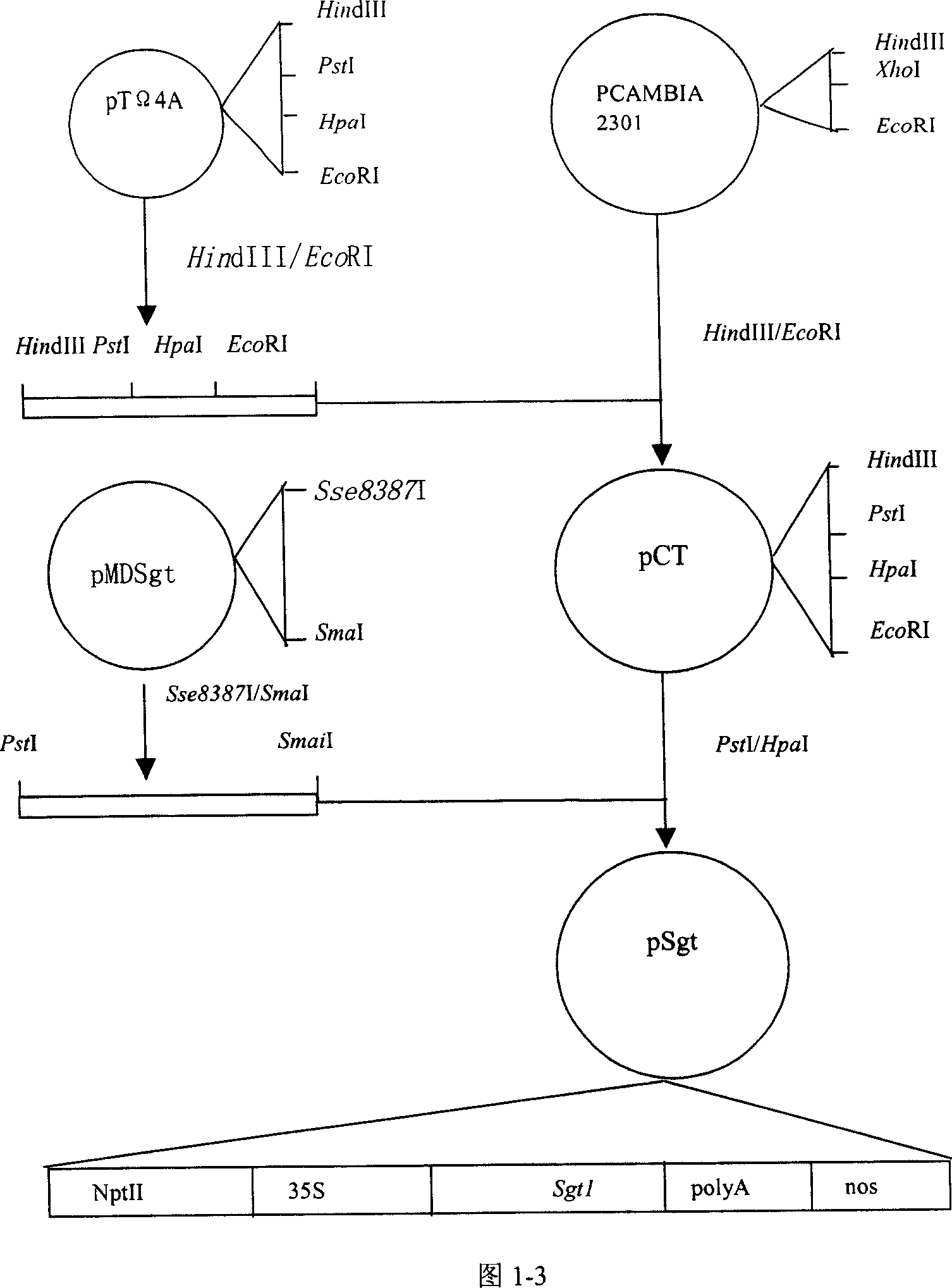
[0111] Embodiment 2: the construction of GbRar1, GbSgt1, GbRac1 gene plant expression vector
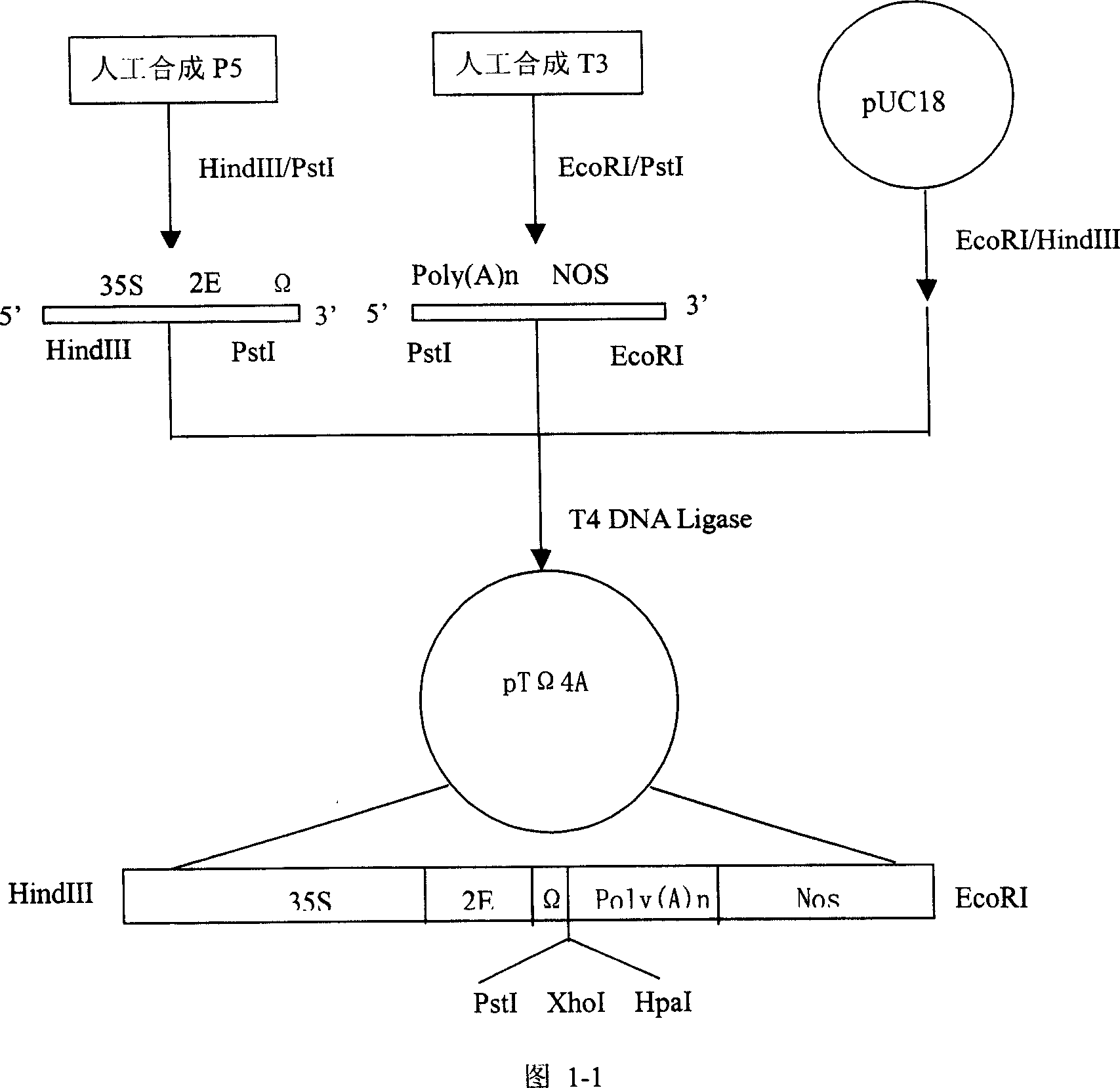
[0112] 2.1 Construction of target gene expression cassette
[0113] As mentioned above, the gene expression cassette constructed in this example includes the following gene expression regulatory elements: 35S promoter at the 5' end, doubled enhancer, Ω sequence and Kozak sequence, concatenated termination sequence and cutting sequence at the 3' end , NOS terminator. DNA fragment P5 including 35S promoter, doubled enhancer, Ω sequence and Kozak sequence and DNA fragment T3 including concatenated termination sequence, cleavage sequence and NOS terminator were synthesized by artificial synthesis methods well known in the art. For the convenience of construction, HindIII and PstI restriction sites were introduced at the 5' and 3' ends of P5, respectively, and PstI and EcoRI restriction sites were introduced at the 5' and 3' ends of T3, respectively. Digest P5 with HindIII / PstI, treat T...
Embodiment 3
[0121] Example 3: Introduction and Identification of GbRar1, GbSgt1, GbRac1 Gene Plant Expression Vectors in Model Plant Nicotiana tabacum
[0122] 3.1 Preparation of LBA4404 Competent Cells
[0123] Pick a single colony of LBA4404 (CLONTECH company, catalog number #6027-1) and inoculate it in 5ml YEB (containing streptomycin 100μg / ml, sucrose 5g+ peptone 5g+ beef extract 5g+ yeast extract 1g+MgSO4.7H2O 0.049g / L , pH7.2), 28°C, 250rpm for overnight cultivation. Take 2ml colony and add it to 50ml YEB medium, continue to cultivate to OD 600 The value is about 0.6. Transfer the bacterial solution to a sterile centrifuge tube, ice bath for 30 minutes, centrifuge at 5000rpm for 5 minutes, and use 2ml of 20mM CaCl 2 Resuspend the bacteria, and distribute 200 μl per tube into sterile small centrifuge tubes.
[0124] 3.2 Transfer of recombinant plasmids into LBA4404 cells
[0125] Add 2 μg of recombinant plasmids pCRar, pCSgt, and pCRac DNA to 200 μl LBA4404 competent cells, put ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com