Gibberella gene engineering bacterium and its preparation and application
A technology of genetically engineered bacteria and gibberellin, applied in the field of genetic engineering, can solve the problems of time-consuming and labor-intensive results, little potential, and unclear goals, and achieve the effects of high efficiency, strong ability, and clear breeding goals
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
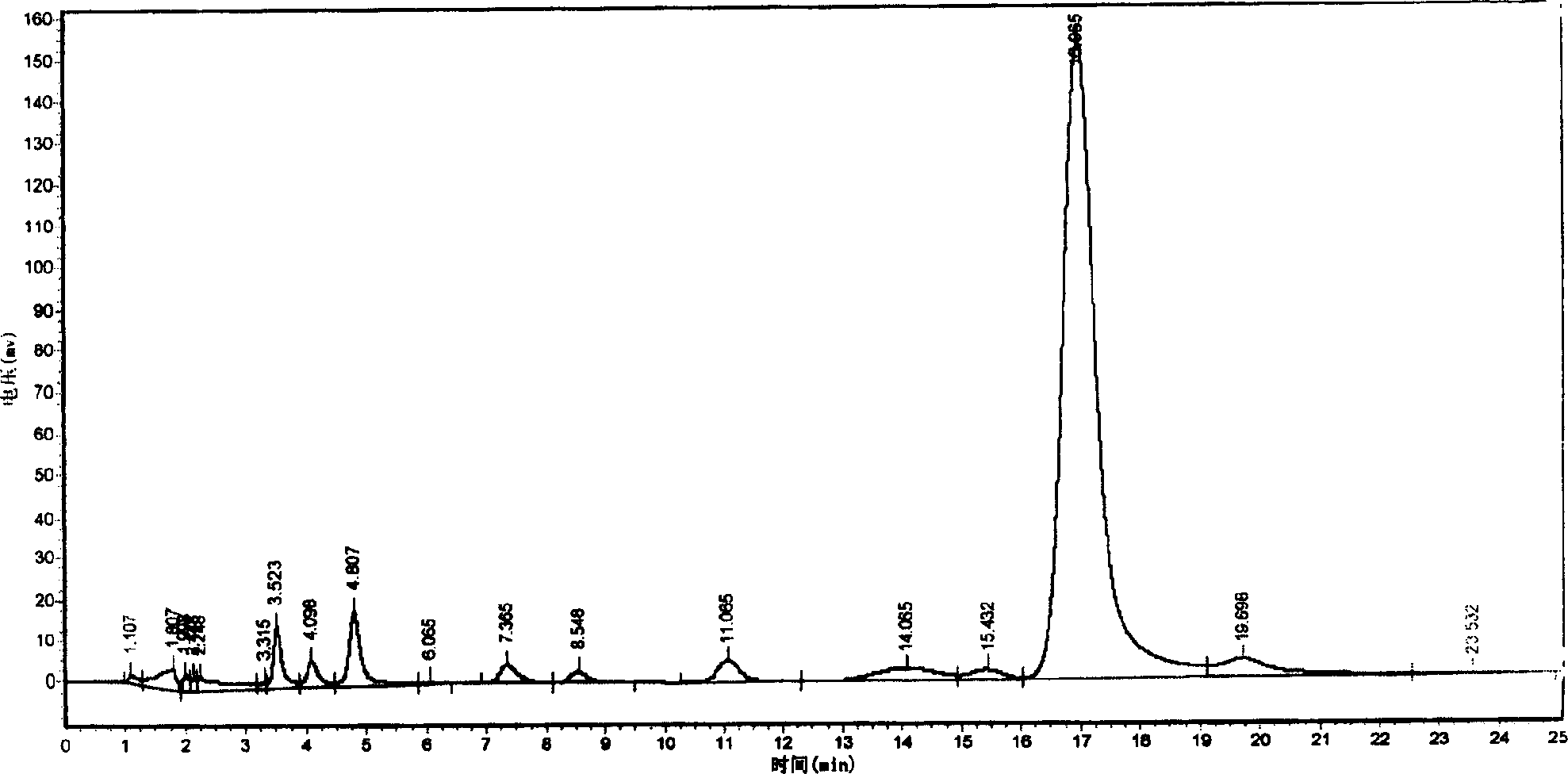
Examples
Embodiment 1
[0054] Embodiment 1: Construction of overexpression vector
[0055] step:
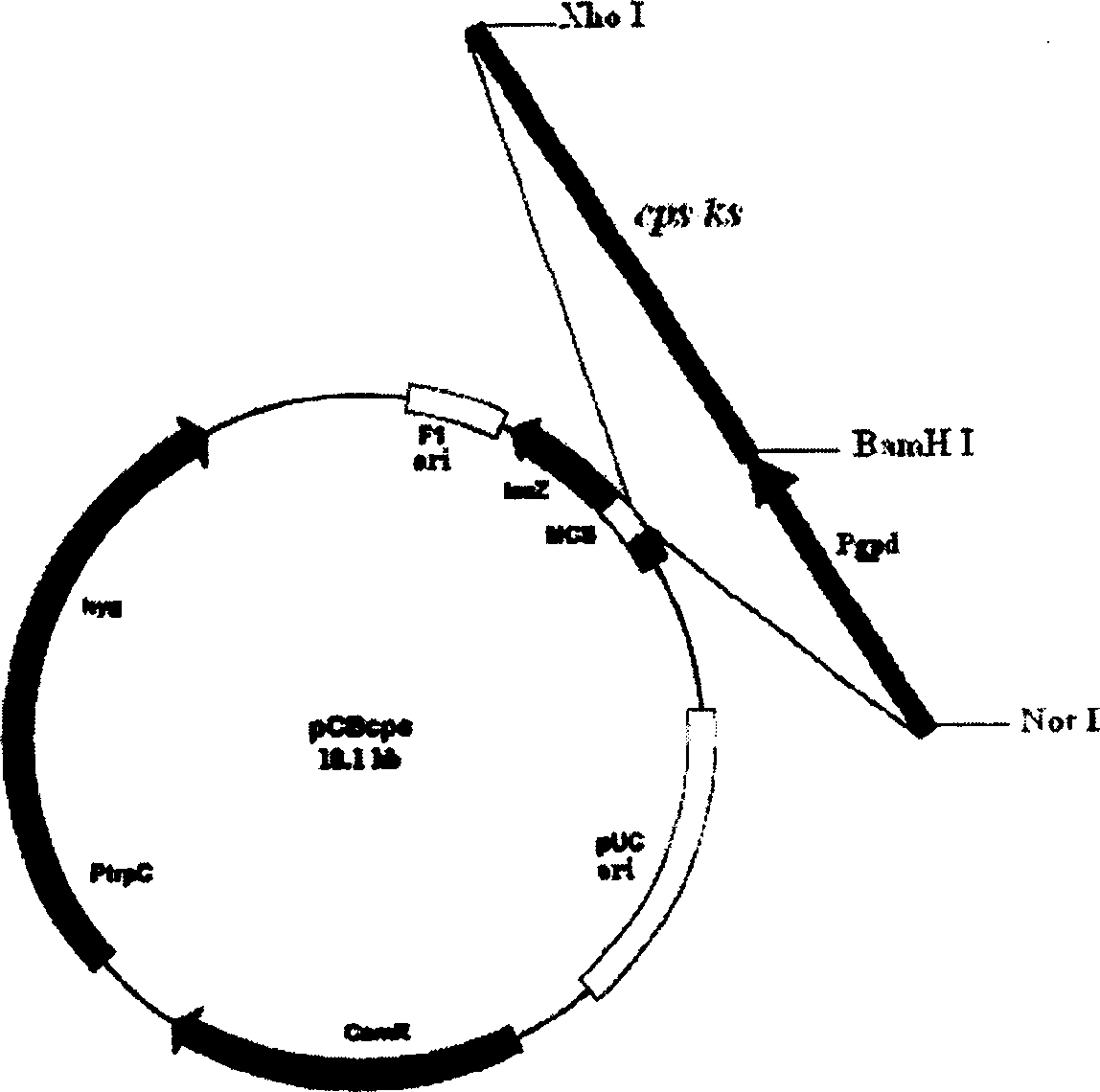
[0056] (1) The strong promoter PgPd was amplified by PCR from the plasmid pAN7-1 (Punt et al.1987Gene56:117-124), and cloned into the vector pCB1004 (Carroll et al.1994Fungal Genet. Newsl.41:22) to form the recombinant plasmid pCB1004Pgpd. The strong promoter Pgpd can also be derived from any other DNA containing this sequence or synthesized directly. In addition to Pgpd, promoters constitutively expressed in filamentous fungi such as PtrpC can also be used. In addition to pCB1004, plasmids used to construct cps / ks gene overexpression vectors can also be expressed in filamentous fungi, such as pCSN43, pCSN44, pCB1004, pCB1003, pSH75, pMYX100, pMSN1 and pAN26.
[0057] (2) Design primers (forward primer: GG) according to the sequence of the gibberellin synthase gene cps / ks (GeneBank accession number: AJ810802) of G. GGATCC GCCGTACGTATGAAACACACCT, the underlined sequence is the cutting site of restr...
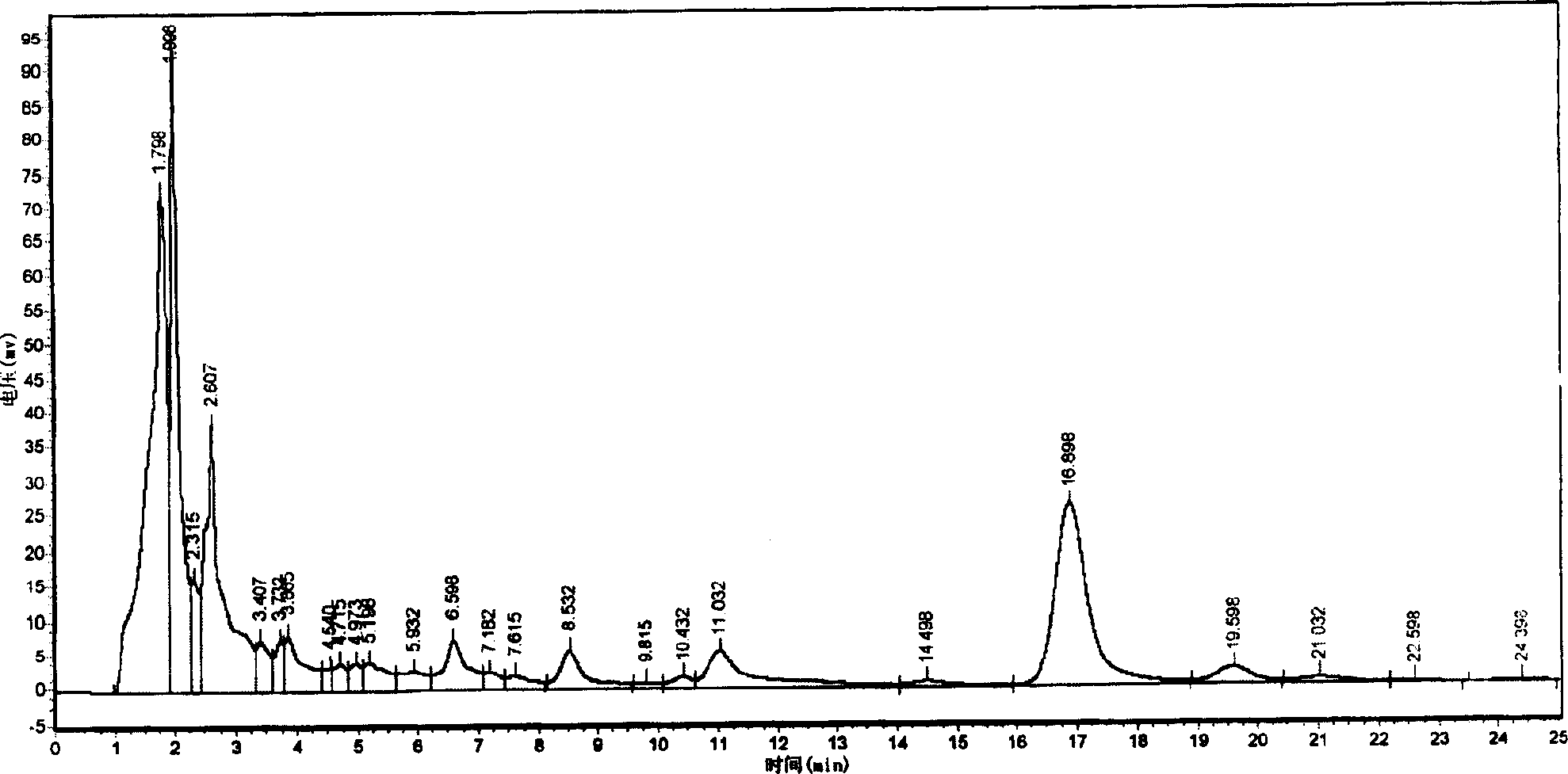
Embodiment 2
[0059] Embodiment 2: Construction of overexpression vector
[0060] step:
[0061] (1) The strong promoter PtrpC was amplified by PCR from the plasmid PZPnat1, and cloned into the corresponding site of the vector pBARKS1 by the restriction enzyme XbaI / BamHI to form the recombinant plasmid pBARtrpC.
[0062] (2) Design primers (forward primer: GG) according to the sequence of the gibberellin synthase gene cps / ks (GeneBank accession number: AJ810802) of G. GGATCC GCCGTACGTATGAAACACACCT, the underlined sequence is the cutting site of restriction enzyme BamH I; reverse primer: CC CTCGAG TACATATCAAA TTACCAGCT, the underlined sequence is the cutting point of restriction enzyme Xho I), and the sequence of the coding region was amplified by PCR technique. PCR reaction conditions: denaturation at 94°C for 5 min; followed by 32 cycles with parameters of 94°C for 1 min; 55°C for 30 sec and 72°C for 2 min; and finally extension at 72°C for 5 min.
[0063] (3) After the PCR product wa...
Embodiment 3
[0064] Embodiment 3: the acquisition of engineering bacteria
[0065] step:
[0066] (1) Inoculate Gibberella fujikura onto a PDA medium plate and cultivate at 24°C for 10 days;
[0067] (2) The spores were washed with sterile water, transferred to 100 mL of YpSs liquid medium, and cultured with shaking at 24°C and 200 rpm for 36 hours;
[0068] (3) Centrifuge at 4000rpm for 5 minutes after the culture solution is filtered to obtain spore precipitation, and wash three times with sterile deionized water;
[0069] (4) Suspend the spores with 200 μl of 0.1M lithium acetate solution;
[0070] (5) Take 5 μl of the plasmid pCBcps obtained in Example 1 (concentration 2 μg / μl), add it to the spore suspension, then add spermidine to make the final concentration 4 mM, and incubate at 4° C. for 30 minutes;
[0071] (6) Add 150 μl of 0.1 M lithium acetate solution of 40% PEG8000, and continue to incubate at 4° C. for 1 hour;
[0072] (7) Heat shock in a water bath at 37°C for 5 minute...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com