Asymmetrical PCR amplification method, dedicated primer and use thereof
An asymmetric, primer pair technology, applied in asymmetric PCR amplification and its application fields, can solve the problems of ineffectiveness, low amplification efficiency, unbalanced amplification of target molecules, etc., to reduce the possibility of interaction, Increase the amplification efficiency and the effect of the same amplification efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0044] Example 1. Single-plex asymmetric PCR amplification and its application in gene chip identification of Gram-positive bacteria
[0045] 1. Gene-specific primers and various oligonucleotide probes for PCR amplification
[0046] The primers and probes were synthesized by Shanghai Boya Biotechnology Company. The TAMR fluorescent labeling of the 5′ end of some primers and the modification of the 5′ end amino group of the probe were also completed by Shanghai Boya Biotechnology Company.
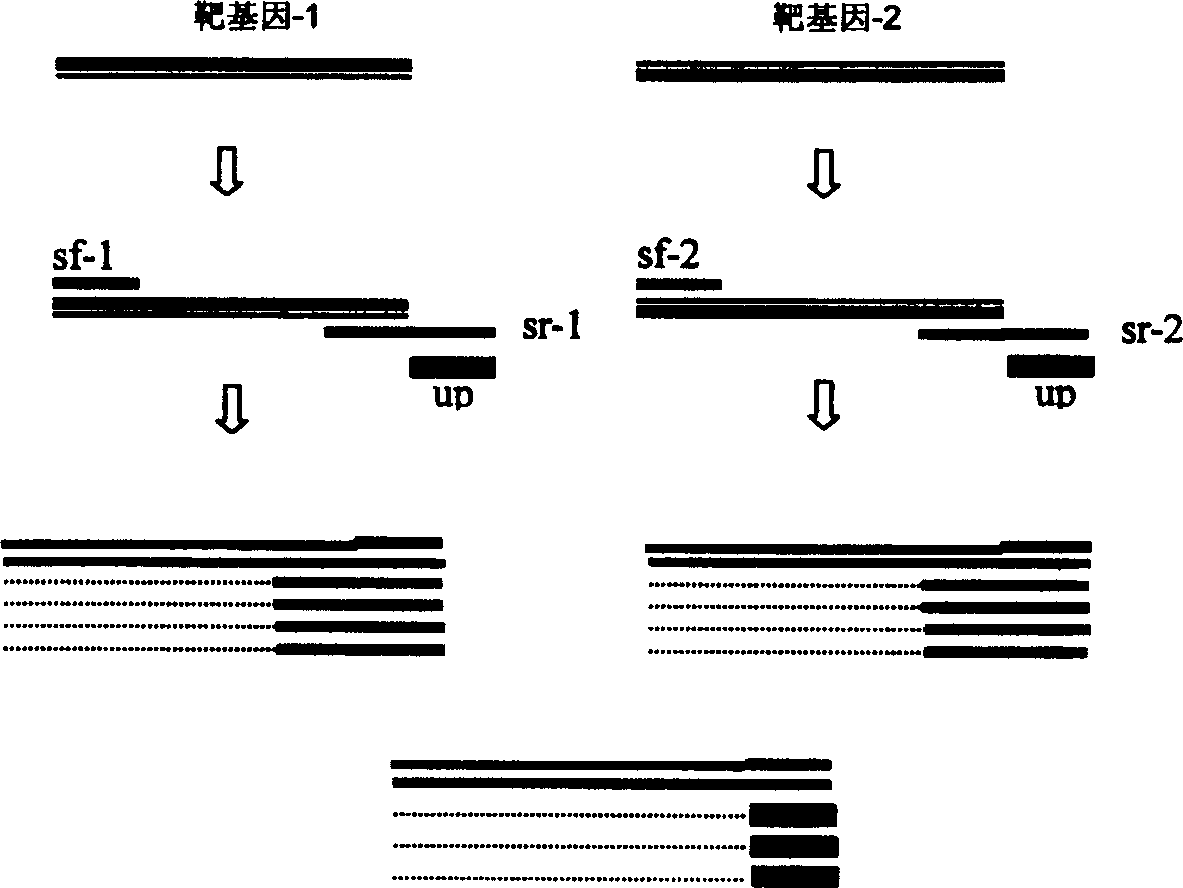
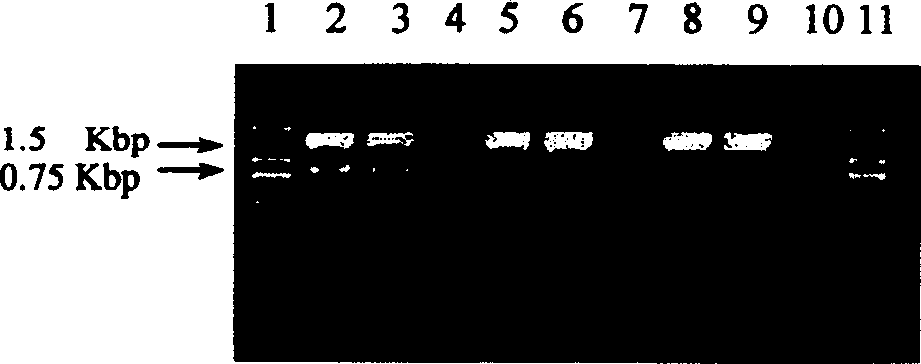
[0047] The target sequence of the gene-specific primers is the bacterial 16S rRNA gene, and the amplified fragment is about 1.5 kb. The types of primers and their nucleotide sequences are shown in Table 1.
[0048] Primer number
Kind of primer
Primer sequence (5′-3′)
PMB-0408047
PMB-0201034
PMB-0201002
PMB-0201042
PMB-0201041
Universal primer
Untailed gene-specific upstream primer
Untailed gene-specific downstream primers
Tailed gene-specific upstream pr...
Embodiment 2
[0081] Example 2. Multiple asymmetric PCR amplification and gene chip detection of bacterial drug resistance genes
[0082] 1. Multiple asymmetric PCR primers and oligonucleotide probes
[0083] The primers and probes used were all synthesized by Shanghai Boya Biotechnology Company. The TAMRA fluorescent labeling of the 5′ end of some primers and the modification of the 5′ end amino group of the probe were also completed by Shanghai Boya Biotechnology Company.
[0084] The types, sequence information, target genes, and amplified fragment lengths of gene-specific primers are shown in Table 7. Two probes were designed for each target gene, and their sequence information is shown in Table 8. Among them, the target genes tetK and tetM are tetracycline resistance genes of Gram-positive bacteria, and the target genes ermA and ermC are Gram-positive bacteria macrolide-lincomycin-streptomycin B resistance genes, 23S A general bacterial sequence of the rRNA gene is used as an internal cont...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com