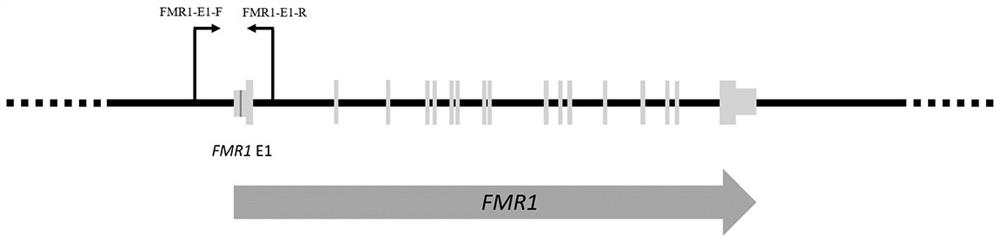
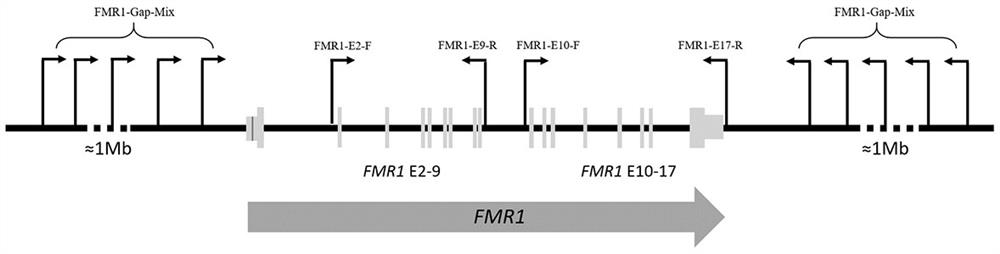
Method and kit for detecting fragile X syndrome mutation
A kit and reagent technology, applied in the field of kits suitable for this method, can solve the problems of omission, inability to directly and accurately determine the number of CGG repeats, inability to detect mutation types at the same time, etc., and achieve detection false detection and missed detection rate. Low, eliminates the influence of noise, and has a wide detection range
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0125] Example 1: Amplify and construct PacBio sequencing library using the PCR method involved in the present invention
[0126] Step 1: Long Fragment and High GC PCR Amplification
[0127] Prepare the reaction system according to Table 1 (long fragment PCR) and Table 2 (high GC PCR) below to amplify peripheral blood, dried blood spot and genomic DNA samples:
[0128]
[0129]
[0130] On the PCR machine, perform pre-amplification according to the conditions shown in Table 3 below:
[0131]
[0132] After the amplification was completed, 5 ul of each sample was taken and tested on a 1% DNA gel. The results were as follows Figure 2A and Figure 2B As shown, using different samples as templates, different fragments of the FMR1 gene can be effectively amplified; at the same time, the amplified products were put into a centrifuge, 10,000 rpm, and centrifuged for 20 min. After centrifugation, it was placed horizontally, and 4 μL of supernatant was added to a new tube ...
Embodiment 2
[0139] Example 2: Construction of PacBio sequencing library using the PCR method involved in the present invention
[0140] Step 1: Long Fragment and High GC PCR Amplification
[0141] Prepare the reaction system according to Table 5 (long fragment PCR) and Table 6 (high GC PCR) to amplify peripheral blood samples with different types of FXS-related gene mutations:
[0142]
[0143]
[0144] On the PCR machine, perform pre-amplification according to the conditions shown in Table 7 below:
[0145]
[0146] After the amplification was completed, the amplified product was placed in a centrifuge at 10,000 rpm for 20 min. After centrifugation, it was placed horizontally, and 4 μL of supernatant was added to a new tube.
[0147] Step 2: Construct PacBio Sequencing Libraries
[0148] The reaction system was prepared according to Table 8 below:
[0149]
[0150] On a PCR machine, perform the reaction as follows: 37ºC for 20 min; 25ºC for 15 min; 65ºC for 10 min. After...
Embodiment 3
[0153]Example 3: Construction of PacBio sequencing library using the PCR method involved in the present invention
[0154] Step 1: Long Fragment and High GC PCR Amplification
[0155] Prepare the reaction system according to Table 9 (long fragment PCR) and Table 10 (high GC PCR) to amplify peripheral blood samples with different types of FXS-related gene mutations:
[0156]
[0157]
[0158] On the PCR machine, perform pre-amplification according to the conditions shown in Table 11 below:
[0159]
[0160] After the amplification was completed, the amplified product was placed in a centrifuge at 10,000 rpm for 20 min. After centrifugation, it was placed horizontally, and 4 μL of supernatant was added to a new tube.
[0161] Step 2: Construct PacBio Sequencing Libraries
[0162] The reaction system was prepared according to Table 12 below:
[0163]
[0164] On a PCR machine, perform the reaction as follows: 37ºC for 20 min; 25ºC for 15 min; 65ºC for 10 min. Aft...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com