Application of Ash1p as negative regulatory factor in improvement of protein expression in host cells
A technology of negative regulators and host cells, applied in the fields of molecular biology and bioengineering, can solve problems such as the unreported function of Ash1p transcriptional regulation, and achieve the effect of improving expression intensity and high-efficiency expression
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
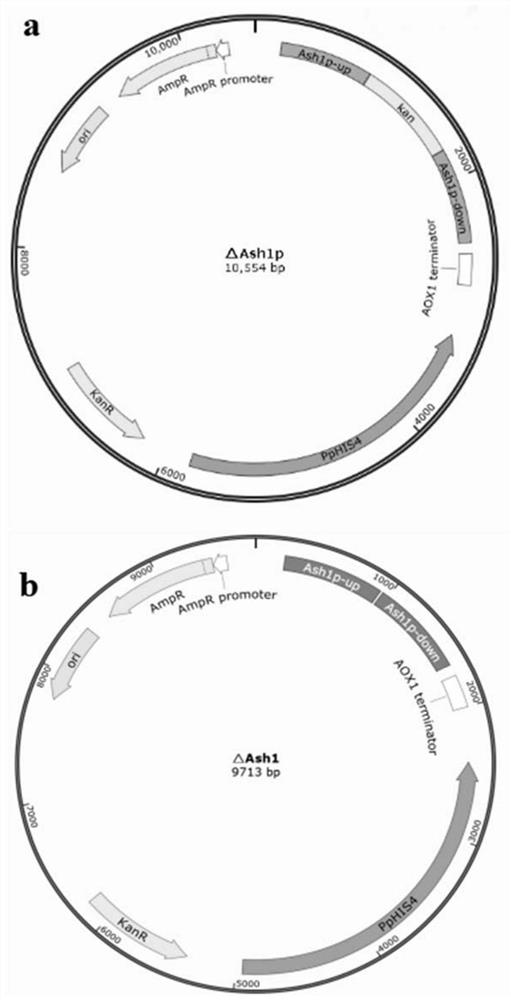
[0021] Example 1: Construction of Kan gene replacement transcription inhibitor Ash1 gene knockout expression cassette 1. Amplification of upstream homology arms
[0022] The present invention takes the genome sequence of Pichia pastoris as a reference, uses software DNAMAN 8 to design and synthesize two oligonucleotide primers, and amplifies the upstream homology arm sequence of Ash1 (SEQ ID NO: 1) by PCR method.
[0023] The two PCR primers are as follows:
[0024] 1-F: ACTTAC GAGCTC GACAGGCAAATCATTCA (SEQ ID NO: 4)
[0025] 1-R: ACTTACGC GGATCC GCGCGTATTTAGCTACAATC (SEQ ID NO: 5)
[0026] The underlined bases are SacI and BamHI restriction endonuclease sites, respectively.
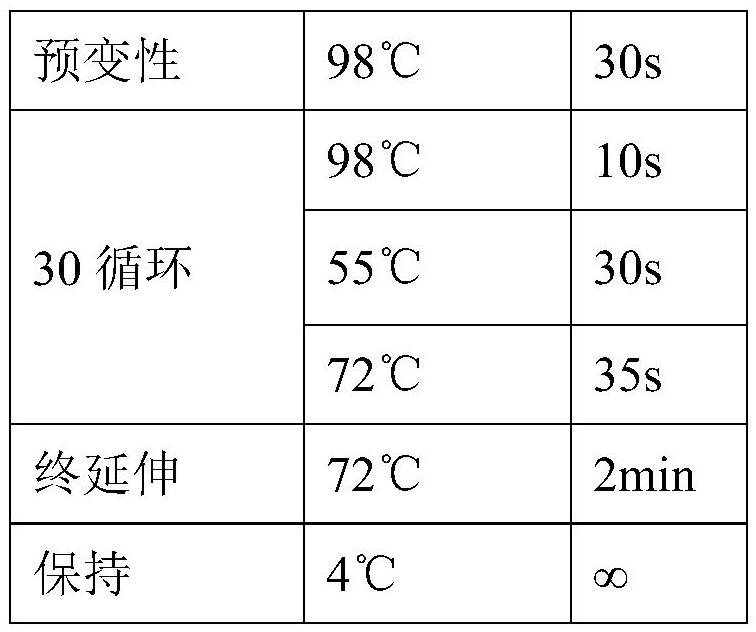
[0027] The PCR reaction system is shown in Table 1 below.
[0028] Table 1:
[0029] component Volume (μL) 2×Q5 Master Mix 12.5 Primer F (10μM) 1.25 Primer R (10μM) 1.25 template DNA 1(100ng) ddH 2 O
9ul total capacity 25ul
[0030] The PC...
Embodiment 2
[0066] Example 2: Construction of expression cassette for complete knockout of transcription repressor Ash1 gene
[0067] As in the method in Example 1, primers 1-F, 1-R and 2-F, 2-R were used to amplify the upper and lower homology arms of the Ash1 gene, and the upstream and downstream homology arm fragments were ligated by the above enzyme cleavage. Methods The ppic3.5k vector was linked to construct ppic3.5k-(upstream homology arm)-(downstream homology arm) knockout expression cassette.
Embodiment 3
[0068] Example 3: Pichia genome Ash1 knockout
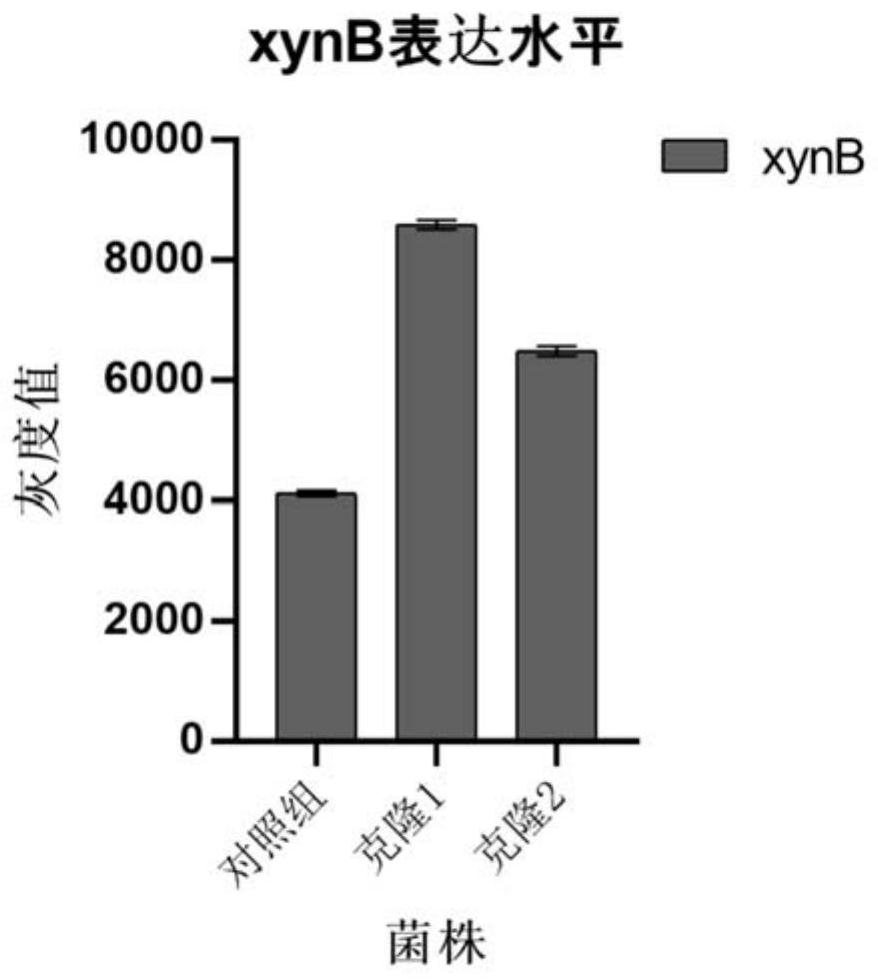
[0069] In order to improve the integration efficiency of the single-copy expression cassette on the chromosome of Pichia pastoris, the knockout expression cassette was linearized with restriction endonucleases Sad and NotI and purified and recovered with the kit. The recipient bacteria in this experiment is Pichia pastoris SMD1168 (recombinant bacteria with xylanase xynB gene inserted after Pgap, EX6), the G418 plate containing 0.3 mg / mL was used after electrotransformation of the knockout expression cassette in Example 1 Screening was performed and genomic PCR identification was performed. Example 2 After electrotransformation of the knockout expression cassette, YPG plate was used for screening, and genomic PCR identification was carried out.
[0070] The results of PCR product sequencing showed that the screened strains were positive clones that successfully knocked out Ash1.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com