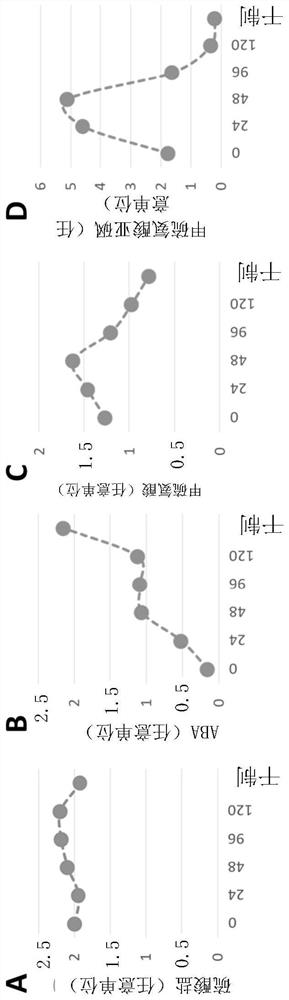
Modulation of sugar and amino acid content (SULTR3) in plants
A plant and plant cell technology, applied in plant genetic improvement, botanical equipment and methods, plant peptides, etc., can solve problems such as limited opportunities for tobacco products and limited types of tobacco
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0215] 6. Preparation, screening and hybridization of modified plants
[0216] NtSULTR3 or NtSULTR3 and NtSUS polynucleotides prepared from individual plants, plant cells or plant material may optionally be combined to expedite screening for mutations in plant populations derived from mutagenized plant tissues, cells or material. One or more subsequent generations of plants, plant cells or plant material can be screened. The size of the optionally pooled cohort depends on the sensitivity of the screening method used.
[0217] After the samples are optionally pooled, they can be subjected to polynucleotide-specific amplification techniques, such as PCR. Any one or more primers or probes specific for the gene or for sequences immediately adjacent to the gene can be used to amplify the sequences within the optionally pooled samples. Suitably, one or more primers or probes are designed to amplify regions of the locus where useful mutations are most likely to occur. Most prefera...
Embodiment 1
[0320] Example 1 - Materials and methods
[0321] DNA extraction and plant genotyping
[0322] Leaf samples were extracted using BioSprint 96 (Qiagen, Hilden, Germany) and the BioSprint 96 DNA Plant Kit (Qiagen, Hilden, Germany). DNA samples were used in TaqMan reactions to determine plant genotypes. Taqman was performed using the ABI PRISM 7900HT Sequence Detection System (Applied Biosystems, Life Technologies, Foster City, CA, USA) and TaqMan Rapid Advanced Master Mix (Applied Biosystems, Foster City, CA, USA).
[0323] Measuring Free Amino Acid Content
[0324] Amino acid content can be measured using various methods known in the art. One such method is Method MP 1471 rev 5 2011, Resana, Italy: Chelab Silliker S.r.l, Mérieux NutriSciences Company. To determine the amino acids in dried plant leaves, the dried leaves were dried at 40°C for 2-3 days after removal of the midrib, if necessary. The tobacco material was then ground to a fine powder (approximately 100 uM) prio...
Embodiment 2
[0327] Analysis of the expression of embodiment 2-NtSULTR gene
[0328] Table 1 shows that NtSULTR3;3-T is expressed throughout plant tissues, especially in petals. Interestingly, copies of NtSULTR3;3-S are not expressed in N. virginica, but in some other tobaccos, such as TN90. Apparently, the NtSULTR3;3-S genomic sequence was not identified in the Virginia tobacco genome or was altered in Virginia tobacco and dark tobacco (Sierro et al. (2014) Nat Commun. May 8;5:3833, see Table 3 and 4). The NtSULTR3;3-S genome and polypeptide sequences were deduced from the TN90 sequencing library. Other SULTR3 genes expressed in petals were NtSULTR3;1A-S, NtSULTR3;1A-T and NtSULTR3;1B-S. NtSULTR3;4A-T is apparently more specific for stems. NtSULTR3;2-S is expressed in sepals and roots. Interestingly, several SULTR3 genes were not expressed or weakly expressed in green leaves, namely, NtSULTR3; 1A-S, NtSULTR3; 1A-T, NtSULTR3; 3-S, NtSULTR3; 4A-S, NtSULTR3; 4B-S, NtSULTR3; 4B-T, NtSUL...
PUM
| Property | Measurement | Unit |
|---|---|---|
| size | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com