Method for predicting ligand-protein interaction based on quantum calculation
A protein interaction and quantum computing technology, which is applied in quantum computers, calculations, molecular design, etc., can solve problems such as difficult predictions, large volumes of proteins with large calculations, and complex spatial structures.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
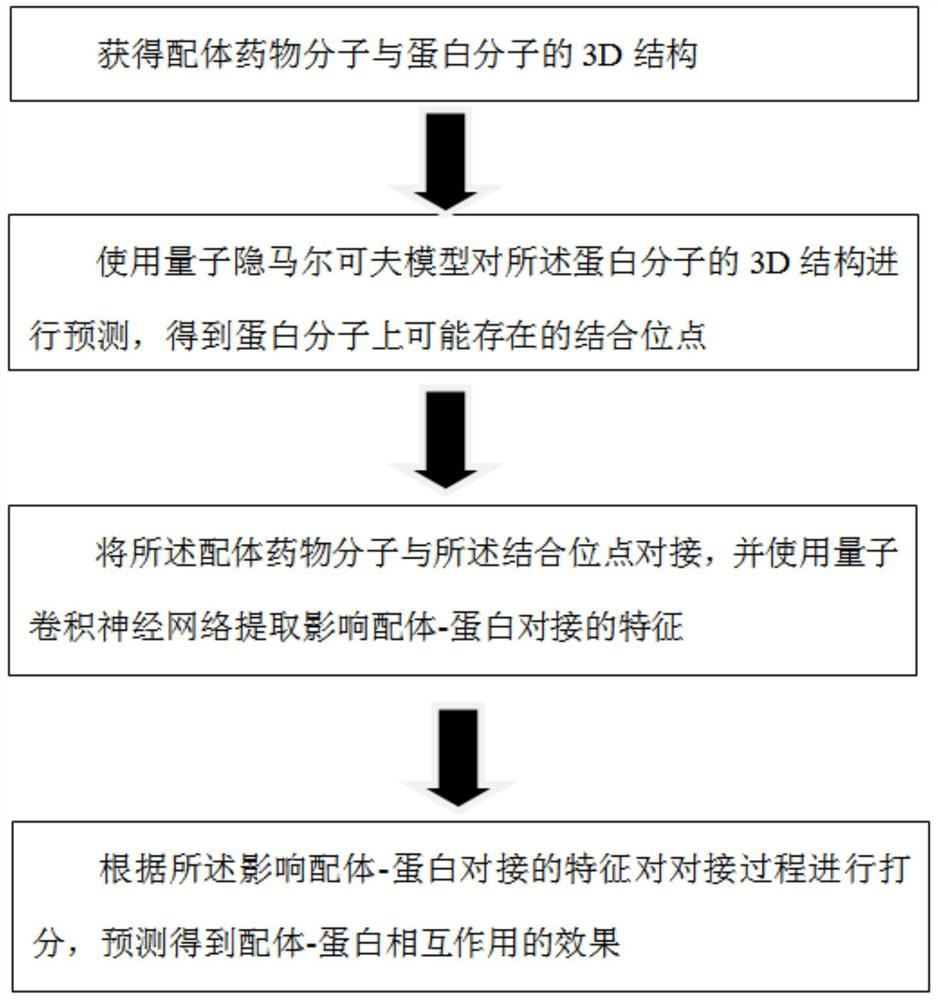
[0061] Provide a prediction method based on quantum computing for ligand-protein interactions, such as figure 1 As shown, the method includes:
[0062] Obtain the 3D structures of ligand drug molecules and protein molecules;
[0063] Use the quantum hidden Markov model to predict the 3D structure of the protein molecule, and obtain the possible binding sites on the protein molecule;
[0064] Docking the ligand drug molecule with the binding site, and using a quantum convolutional neural network to extract features that affect ligand-protein docking;
[0065] The docking process is scored according to the characteristics affecting ligand-protein docking, and the effect of ligand-protein interaction is predicted.
[0066] Specifically, the 3D structure of the drug molecule and the 3D structure of the protein molecule are respectively obtained by methods such as crystallography or spectroscopy for prediction training. Then use the quantum hidden Markov model combined with the ...
Embodiment 2
[0071] Based on Example 1, a method for predicting ligand-protein interaction based on quantum computing is provided, wherein the obtaining of the 3D structure of the ligand drug molecule and the protein molecule includes:
[0072] Obtain the three-dimensional crystal structure of protein molecules by means of X-ray crystallography or NMR spectroscopy;
[0073] Obtain the 3D structures of a large number of ligand drug molecules with the help of a large database of small molecule 3D structures.
[0074] Further, said obtaining the 3D structure of the ligand drug molecule and the protein molecule also includes:
[0075] Repair the errors generated during the protein analysis process, and perform hydrogenation protonation on the three-dimensional crystal structure of the protein molecule before docking, mark the local electrical properties, and wait for the next step;
[0076] Energy minimization is performed on the 3D structure of the ligand drug molecule.
[0077] Further, as...
Embodiment 3
[0100] This embodiment has the same inventive concept as Embodiment 1. On the basis of Embodiment 1, a storage medium is provided on which computer instructions are stored. When the computer instructions are running, a quantum computing-based Steps in a predictive method for ligand-protein interactions.
[0101] Based on this understanding, the technical solution of this embodiment is essentially or the part that contributes to the prior art or the part of the technical solution can be embodied in the form of a software product, and the computer software product is stored in a storage medium. Several instructions are included to make a computer device (which may be a personal computer, a server, or a network device, etc.) execute all or part of the steps of the methods in various embodiments of the present invention. The aforementioned storage medium includes: U disk, mobile hard disk, read-only memory (Read-Only Memory, ROM), random access memory (Random Access Memory, RAM), ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com