Mutant RNase R as well as preparation method and application thereof
A mutation-type, site-directed mutation technology, applied in the field of molecular biology, can solve the problems of increasing production costs and loss, and achieve the effect of high protein expression
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
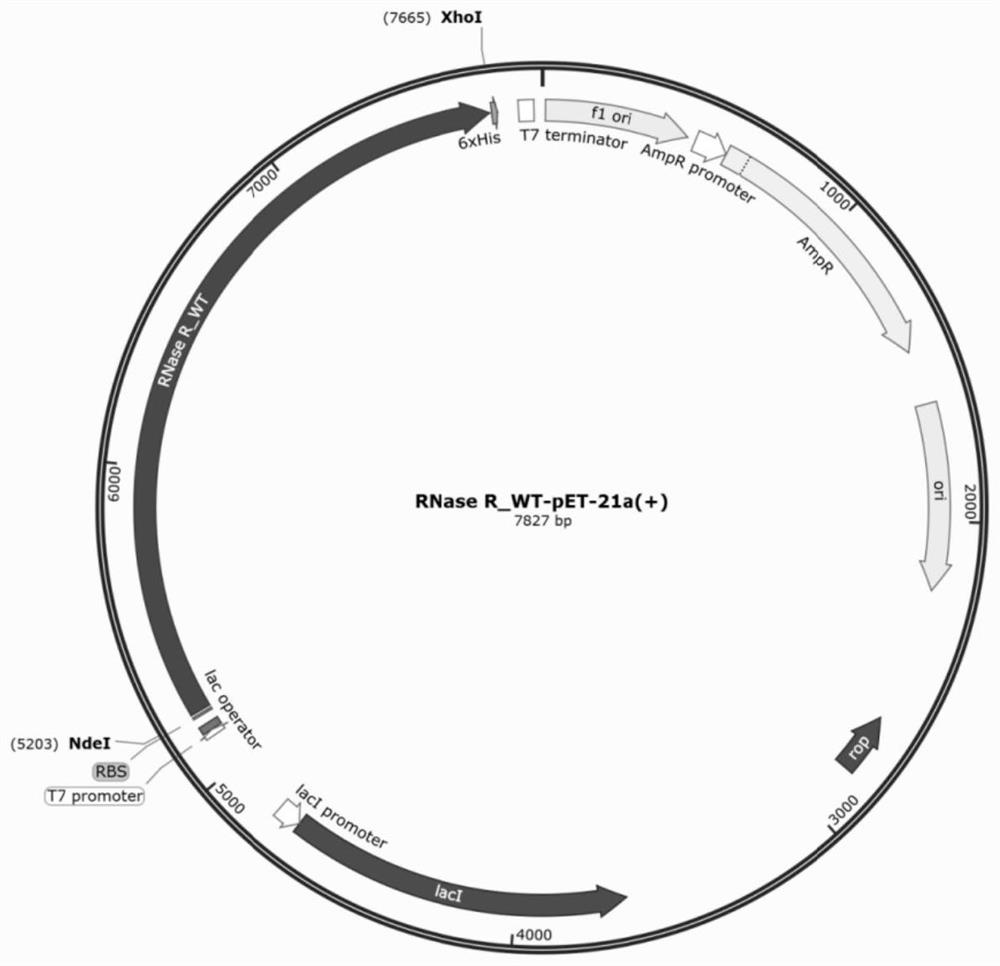
[0037] The structure of embodiment 1 mutant RNase R expression vector
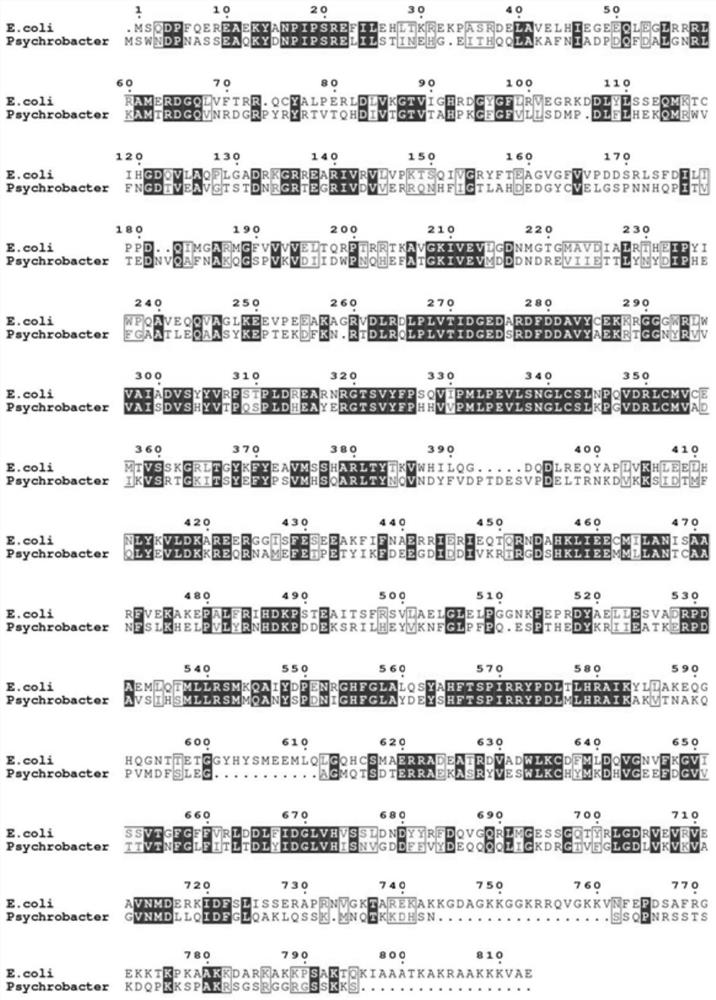
[0038] The wild-type RNase R (amino acid sequence shown in SEQ.NO.1) of Escherichia coli origin and the RNase R (amino acid sequence shown in SEQ.NO.3) of a salt-tolerant Psychrobacter sp.strain ANT206 origin were carried out amino acid Sequence alignment (alignment results such as figure 1 shown). Determine the amino acid residues that may have an important impact on salt tolerance, and perform truncation mutations on them. The mutated RNase R is named RNase R_△M8, and its amino acid sequence is shown in SEQ.NO.5.
[0039] Primers were designed according to the mutation points, and the primer sequences were as follows:
[0040] RNase R-F: TAACTTTAAGAAGGAGATATACATATGCATCATCATCATCATCATTCACAAG (SEQ. ID NO. 9);
[0041] RNase R_△M8-R: TGCCGGTTTCAGTGGTGTTGCCCTG (SEQ.NO.10);
[0042] RNaseR_ΔM8-F: GCAACACCACTGAAACCGGCATGCTGCAACTGGGTCAGCAC (SEQ. NO.11);
[0043] RNase R-R: TCAGTGGTGGTGGTGGTGGTGCTCGAGTCACTCT...
Embodiment 2
[0049] Example 2 protein expression
[0050] The expression strain RNase R_ΔM8 (BL21(DE3)) obtained in Example 1 was inoculated into 5 mL of LA culture medium, and placed in a shaker at 37° C. at 200 rpm for overnight shaking culture.
[0051] On the next day, inoculate 5 mL of the overnight culture into a new 500 mL LA culture medium, culture on a shaker at 37°C and 200 rpm for 3 hours (OD value about 0.5), add 500 μL IPTG (1M) to the culture medium (final concentration 1 mM), 37 ℃, 200rpm shaker to continue culturing for 3h. The cells were collected by centrifugation at 10000 g for 5 min, and washed once with 5 mL of sterile PBS.
Embodiment 3
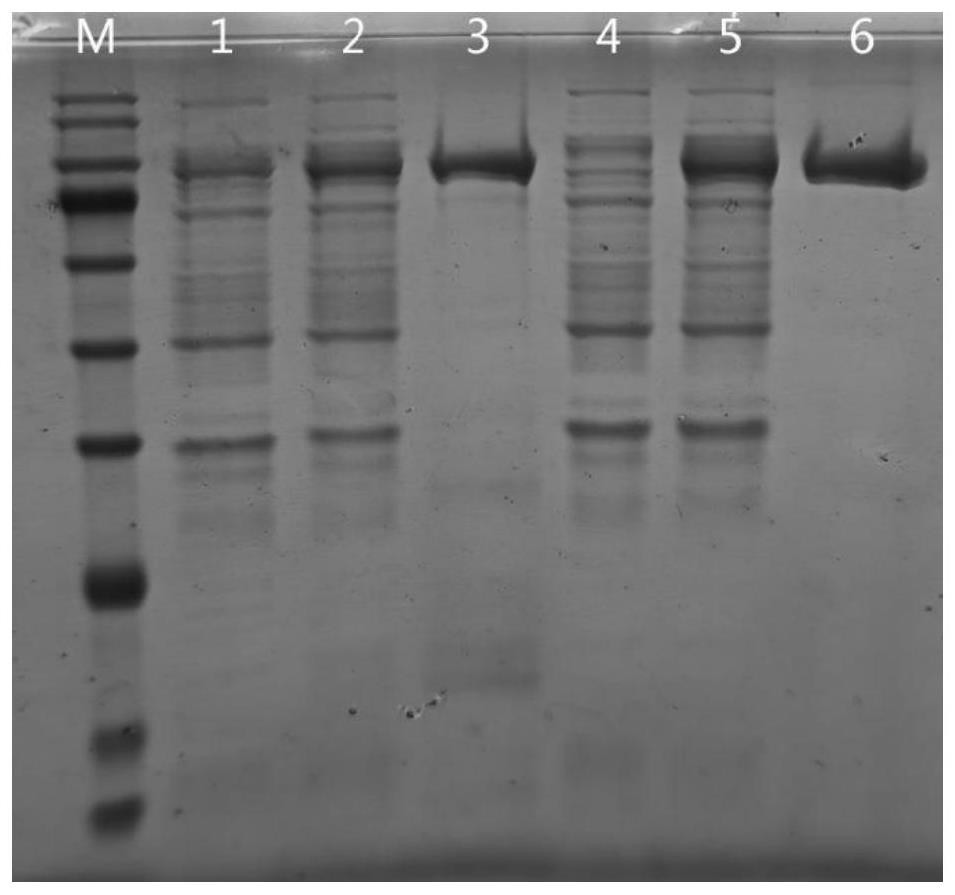
[0052] Example 3 protein purification
[0053] Add 40mL balance washing buffer (50mM phosphate, 500mMNaCl, 20mM imidazole, 0.05% Tween 20, 10% Glycerol, pH 8.0) to the cells collected in Example 2, vortex until the cells are fully resuspended, and centrifuge The tube was fixed in an ice-water bath, the ultrasonic probe was inserted into the liquid surface 1-2cm below the liquid surface, ultrasonicated until the bacterial liquid was clear and transparent (75% power, ultrasonic 4s and 6s off, total time 10min), centrifuged at 18000g, 4°C for 60min, and the supernatant solution (i.e. RNase R_△M8 protein lysate) was transferred to a new centrifuge tube.
[0054] Protein purification was performed using a protein purification system (Unique AutoPure, Inscinstech):
[0055] Ni-NTA column purification: After flushing the system pipes and Ni-NTA column (BBI, product number C600792, specification 1mL) with DEPC treated water, equilibrate the column with equilibrium washing buffer. Lo...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com