Rice SNP (Single Nucleotide Polymorphism) marker and application thereof
A marker, rice technology, applied in recombinant DNA technology, DNA/RNA fragment, determination/inspection of microorganisms, etc., can solve the problems of quantitative judgment of adulteration, prone to misjudgment, poor accuracy, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0082] Example 1: Materials and methods
[0083] 1. Materials
[0084] Wuyoudao 4-rice seeds, seeds of other rice varieties and commercially available rice samples were all provided by Wilmar (Shanghai) Biotechnology R&D Center Co., Ltd.
[0085] 2. Enzymes and Reagents
[0086] The enzymes were purchased from Kangwei Century Company and Bio-Rad Company, the reagents were purchased from Sinopharm Chemical Reagent Co., Ltd., and the fluorescent quantitative PCR instrument Bio-Rad CFX96; the primers and probes used in the experiment were synthesized by Shanghai Sangon Bioengineering Company .
[0087] 3. Experimental method
[0088] 3.1 DNA extraction from rice seeds (rice samples)
[0089] Grind 20 grams of rice seeds (rice samples) with a grinder, weigh 50 mg of powder into a 2 mL sample lysis tube, add 500 μL of Buffer 1, vortex for 30 seconds, incubate at 52°C for 30 minutes at 1200 rpm; add 500 μL Buffer2, vortex and mix for 30s, centrifuge the mixture for 5min (12000r...
Embodiment 2
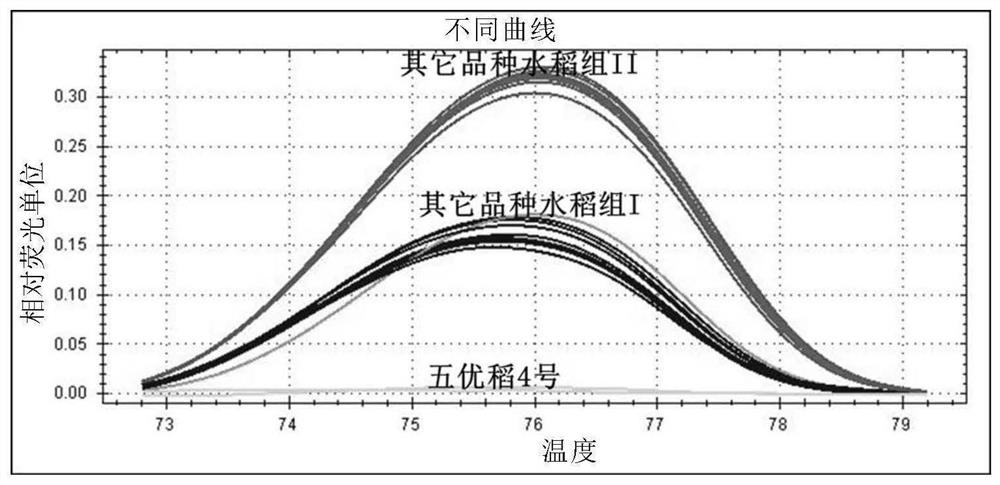
[0112] Embodiment 2: HRM high-resolution melting curve method real-time fluorescent quantitative PCR detection
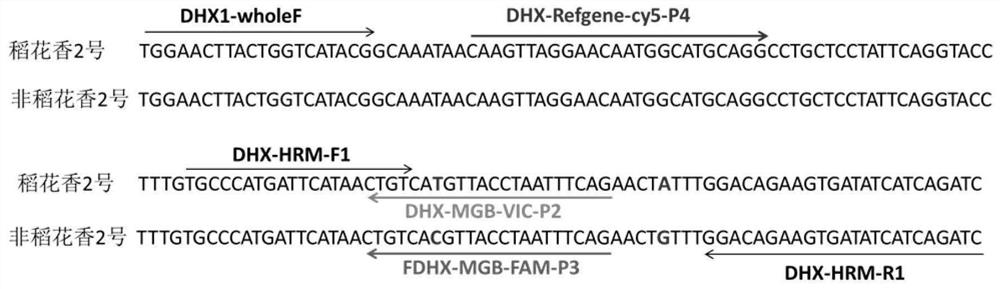
[0113] Using the extracted DNA as a template, the HRM high-resolution melting curve method was used to detect the primer pairs DHX-HRM-F1 and DHX-HRM-R1 for real-time fluorescent quantitative PCR amplification. The PCR reaction system is 20μL, in which SsoAdvancedTM Green Supermix 10μL, DHX-HRM-F1 and DHX-HRM-R1 primers (concentration: 10μM) 0.5μL each, template DNA (concentration: 50-100ng / μL) 2μL, sterile water to make up to 20μL. In the blank control, sterile water was used instead of template DNA. Each reaction was repeated three times, and the PCR amplification program used a two-step method: pre-denaturation at 95°C for 3 minutes; denaturation at 95°C for 15 seconds, annealing and extension at 60°C for 1 minute, a total of 45 cycles.
[0114] After the real-time fluorescent PCR amplification is completed, the amplified product is directly applied to Bio-Rad...
Embodiment 3
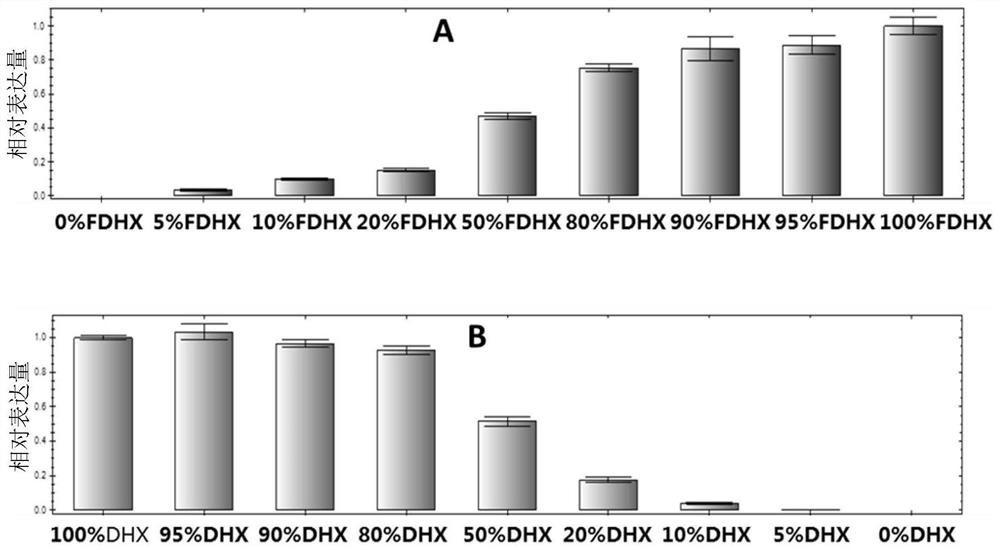
[0116] Embodiment 3: Probe method real-time fluorescence quantitative PCR detects
[0117] Using the extracted DNA as a template, use the Wuyoudao No. 4 probe method to detect the primers. Real-time fluorescent PCR detection was carried out with non-Wuyoudao No. 4 specific probe FDHX-MGB-FAM-P3 and endogenous reference gene DHX-Refgene-cy5-P4 probe. The PCR reaction system was 20 μL, including 10 μL of 2×GoldStar Best MasterMix, 0.8 μL of DHX1-wholeF and DHX-HRM-R1 primers (10 μM concentration), 0.25 μL each of DHX-MGB-VIC-P2 probe (10 μM concentration), FDHX-MGB-FAM-P3 probe (concentration: 10 μM) 0.125 μL each, DHX-Refgene-cy5-P4 probe (concentration: 10 μM) 0.2 μL, template DNA 2 μL, sterile water to make up to 20 μL. In the blank control, sterile water was used instead of template DNA. Each reaction was repeated three times, and the PCR amplification program used a two-step method: pre-denaturation at 95°C for 10 min; denaturation at 95°C for 15 s; annealing and extensio...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com