Method for researching corn southern rust group inheritance and transmission based on SNP (Single Nucleotide Polymorphism)
A technology of southern rust and population genetics, applied in the field of molecular biology, can solve the problems such as the absence of SNP markers of Puccinia multicumulus, and achieve the effect of reducing the use of pesticides and reducing yield loss
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0029] The screening of embodiment 1 restriction endonuclease
[0030] Sampling of Puccinia spp. in Hainan, Guangdong and Guangxi of my country, and bringing the collected samples of Puccinia back to the laboratory. Single spore piles were picked for single spore isolation and culture to obtain single spore lines (strains). For the specific method, refer to the invention patent applied by our research group (a method for single spore propagation of Puccinia multicumulus corn, patent number: ZL201510765142. 1), through multiplication, each monospore line obtains a large number of uredospores. Extract DNA from a single spore. The quality of DNA extraction was detected by 0.8% agarose gel electrophoresis, and DNA was quantified by UV spectrophotometer. Use a combination of restriction endonucleases to digest the DNA. After digestion, take 5ul of each sample for gel run detection. The DNA digestion is required to be complete, that is, there is no main band after the digestion; t...
Embodiment 2
[0035] Example 2 SNP marker mining
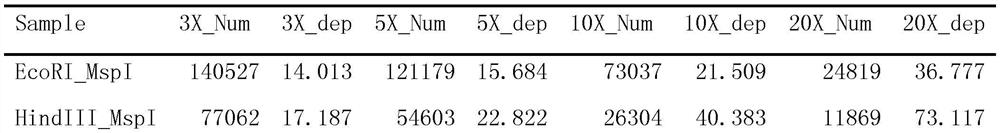
[0036] The DNA of different strains was digested with a combination of EcoRI and MspI restriction endonucleases, and the digested sequence fragments were subjected to pair-end sequencing on the NovaSeq 6000 platform through simplified genome technology, and the length of the sequenced fragments was 150 bp. Using stacks software, the simplified genome sequences of 106 strains collected from Hainan, Guangdong and Guangxi were compared to detect mutation sites and obtain SNP data of different strains.
Embodiment 3
[0037] Example 3 Population Genetics Analysis and Path Inference
[0038] Genetic Diversity Analysis
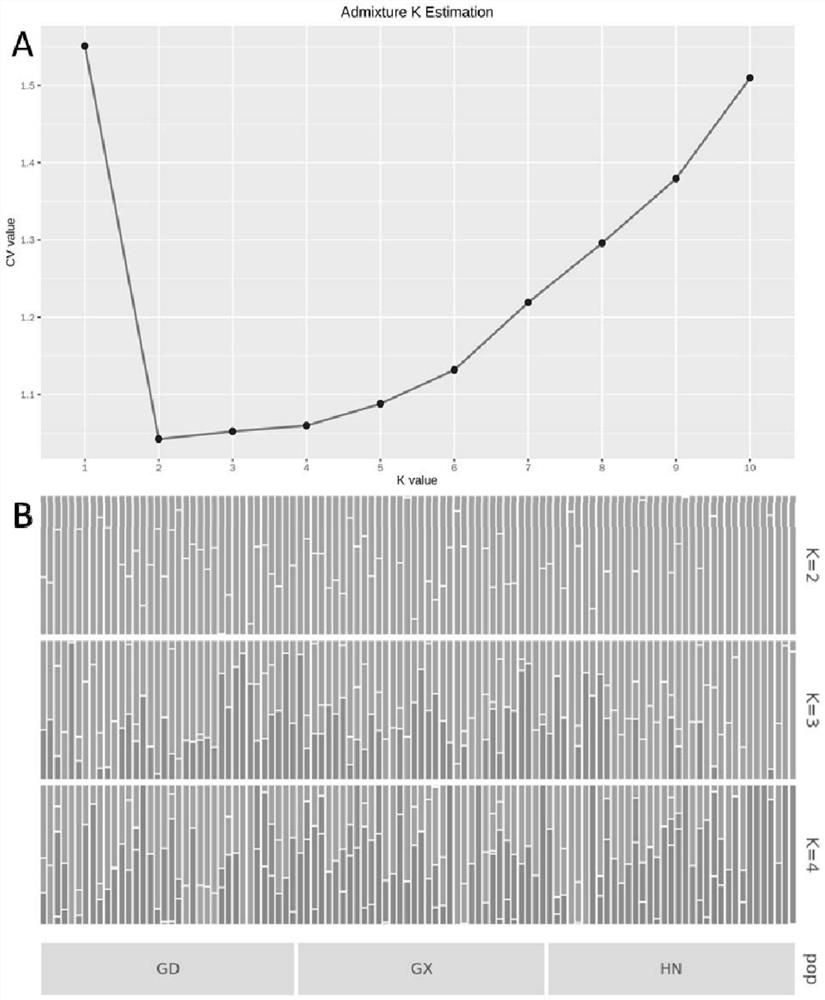
[0039] Based on the SNP data, using the poppr data package of R software (Kamvar et al., 2014), the genetic diversity of a total of 106 Puccinia strains in Hainan, Guangdong and Guangxi was analyzed, including the average individual at each site in the population The number (NumIndv), the average observed heterozygosity of all loci (Obs Het), the average observed homozygosity of all loci (Obs Hom), and the average expected heterozygosity of all stable points in the population (Exp Het ). The strains in Guangdong and Guangxi were found to have higher genetic diversity, while the strains in Hainan had lower genetic diversity (Table 2).
[0040] Table 2 Genetic diversity results of different populations
[0041]
[0042] Note: HN represents the population of Puccinia multicumulus collected from Hainan Province, GX represents the population collected from Guangxi, and GD re...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com