Peach fruit ethylene response factor PpRAP2.12 gene as well as cloning method and application thereof
A pprap2.12, response factor technology, applied in application, genetic engineering, plant genetic improvement, etc., to achieve the effect of reducing sucrose content, increasing decomposition, and increasing VIN activity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
specific Embodiment 1
[0023] Cloning and sequence analysis of the ethylene response factor PpRAP2.12 gene in peach fruit
[0024] 1. Total RNA was extracted from "Yulu" peach fruit, and reversed into cDNA, which was used as a PCR template; the details were as follows: using Promega's EastepTM total RNA extraction kit (Promega, United States) to extract total RNA from peach fruit, and then using HiScript® II Q Select RT Super Mix for qPCR (Vazyme, Nanjing, China) kit was reverse transcribed into cDNA as a template for PCR reaction;
[0025] 2. Use the online website NCBI-PRIMER (https: / / www.ncbi.nlm.nih.gov / tools / primer-blast / ) to design the specific amplification of the CDS region of peach fruit PpRAP2.12 gene (Gene ID: LOC18783672) Primer, upstream primer sequence: 5´-ATGTGTGGAGGTGCTATAATATCCG-3´, downstream primer sequence: 5´-TCAGAAACCTCCCCCAATCAG-3´;
[0026] 3. PCR amplification: The PpRAP2.12 gene amplification product was obtained by PCR amplification. The PCR amplification reaction system ...
specific Embodiment 2
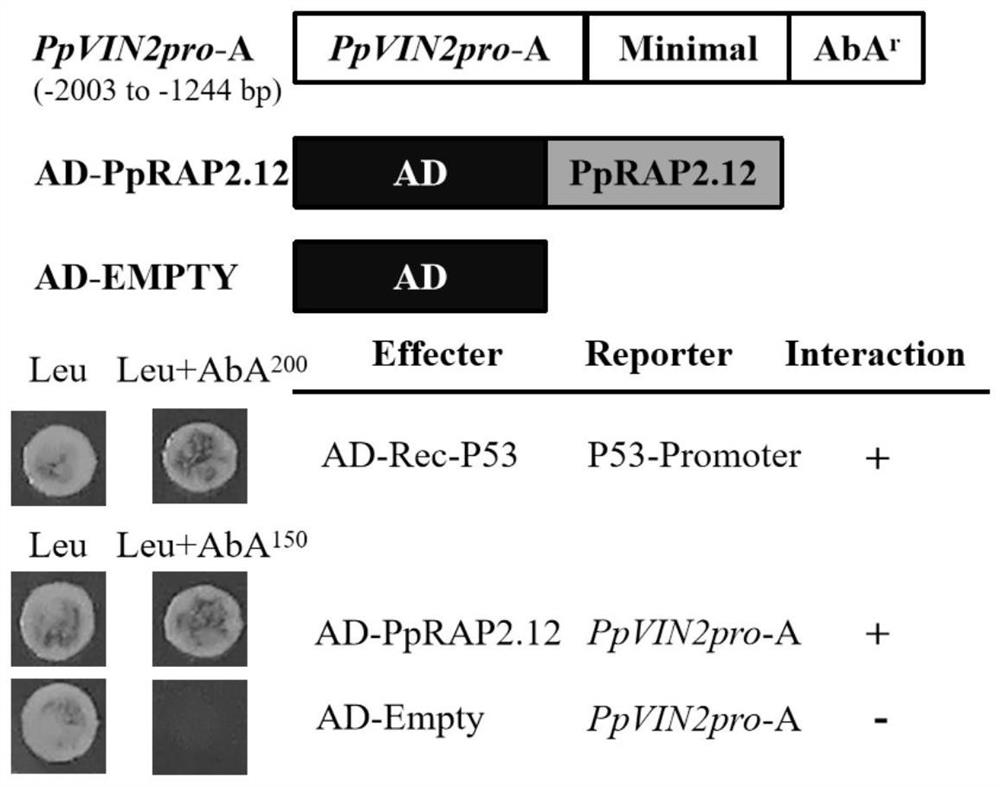
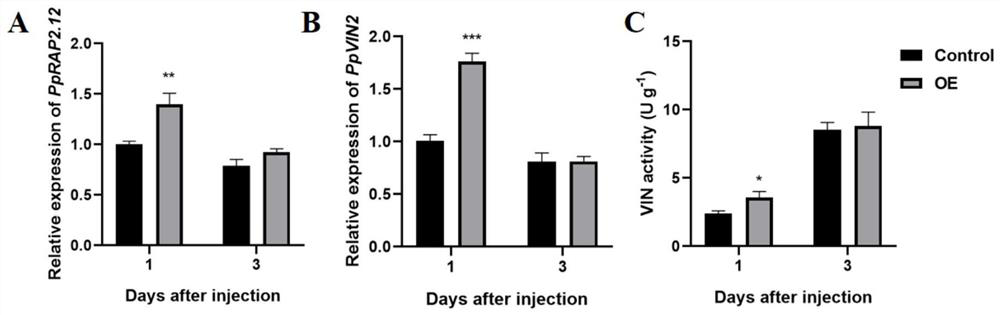
[0032] Confirmation that PpRAP2.12 can bind to the promoter of PpVIN2 gene by yeast one-hybrid system (Y1H)
[0033] 1. Construction and identification of the bait recombinant vector pAbAi-PpVIN2pro-A containing the PpVIN2 promoter truncation (PpVIN2pro-A) and the prey recombinant vector pGADT7-PpRAP2.12
[0034] The PpVIN2 promoter contains abundant transcription factor binding sites. Since the cloned PpVIN2 promoter sequence is relatively long, and the front of the promoter sequence contains a potential binding site GCC-box (ACCGAC) for PpRAP2.12, it is planned to activate PpVIN2 The promoter was truncated, and the front part of the promoter was taken and named PpVIN2pro-A to verify whether it could bind to the PpRAP2.12 transcription factor.
[0035] Use the online website NCBI-PRIMER (https: / / www.ncbi.nlm.nih.gov / tools / primer-blast / ) to design peach fruit PpRAP2.12 gene and PpVIN2 gene promoter truncations (Gene IDs are LOC18783672, LOC18776102) specific amplification pri...
specific Embodiment 3
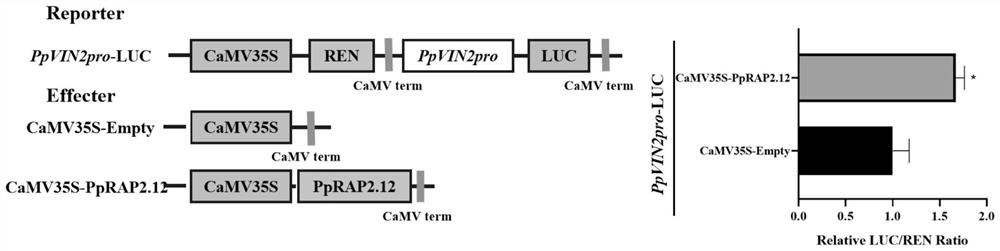
[0046] Validation of the transcriptional activity of PpVIN2 promoted by PpRAP2.12 using dual luciferase assay
[0047] 1. The full-length PpVIN2 promoter was constructed on pGreen Ⅱ 0800-LUC as a reporter vector, and the CDS sequence of PpRAP2.12 was constructed on pGreen Ⅱ 62-sk vector as a control and effector vector.
[0048] Use the online website NCBI-PRIMER (https: / / www.ncbi.nlm.nih.gov / tools / primer-blast / ) to design peach fruit PpRAP2.12 gene and PpVIN2 gene promoters (Gene IDs are LOC18783672 and LOC18776102, respectively). Specific amplification primers, with appropriate restriction sites and protective bases or homologous sequences of the vector at both ends of the primers (Table 2).
[0049]Table 2 Primer sequences for constructing recombinant vectors pGreen Ⅱ 62-sk-PpRAP2.12 and pGreen Ⅱ 0800-PpVIN2pro
[0050]
[0051] Note: The underline indicates the protected base; the bold indicates the restriction site, XhoI, SmaI, BamHI, SmaI
[0052] The reaction syste...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com