Colorectal cancer biomarker and application thereof in diagnosis, prevention, treatment and prognosis
A biomarker and colorectal cancer technology, applied in the field of unsupervised cluster analysis of biomarkers, can solve the problem of unclear subgroup map of ILCs and achieve the effect of survival rate prediction
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
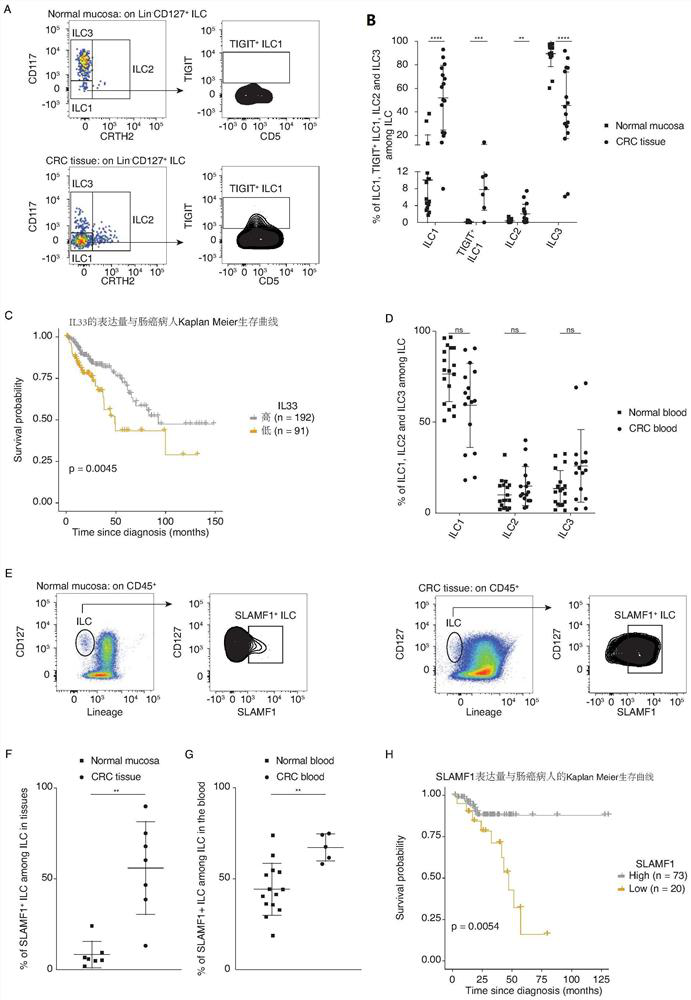
[0147] Example 1 A healthy gut contains ILC1s, IL3Cs and ILCs / NK, but no ILC2s.
[0148] 1. Materials and methods
[0149] (1.1) Collection of clinical samples
[0150] Healthy blood samples for scRNAseq were obtained from individuals undergoing routine colonoscopies, usually in good health and without other relevant medical history, such as inflammatory bowel disease (IBD) or CRC.
[0151] (1.2) Separation of human lymphocytes
[0152] Prepare fresh intestinal tissue immediately after surgery. Adipose tissue and visible blood vessels are removed. Samples were weighed and washed in PBS, then cut into small pieces. Normal tissues were incubated in 10 mL of freshly prepared endothelial lymphocyte solution (PBS containing 5 mM EDTA, 15 mM HEPES, 10% FBS, 1 mM DTT) for 1 hour at 37°C with shaking at 200 rpm. CRC tissue was washed with shaking in 10 mL of freshly prepared PBS containing 65 mM DTT for 15 min at 37 °C. After the incubation, the tissue pieces were washed twice w...
Embodiment 2
[0160] Example 2 The present invention carried out single-cell transcriptome sequencing (scRNAseq) on 58,000 ILCs in blood samples of CRC patients, healthy blood, normal mucosa and CRC tissue samples ( Figure 6 D).
[0161] 1. Materials and methods
[0162] (1.1) Single-cell RNA sequencing
[0163] Purified ILCs were resuspended in PBS containing 0.04% BSA and kept on ice. For cell counting, cell density was adjusted to the concentration suggested by the 10X Genomics v3 kit and used for library construction. The Illumina platform (NovaSeq 6000) of Crystal Energy was used for library sequencing, and the sequencing depth was about 90,000 reads per cell.
[0164] (1.2) Raw sequence alignment, quality control and normalization
[0165] Using FastQC software v0.11.9
[0166] (https: / / www.bioinformatics.babraham.ac.uk / projects / fastqc / ) for quality control of raw sequencing reads. Sequencing data in bcl files were converted to FASTQ format using bcl2fastq2 conversion software ...
Embodiment 3
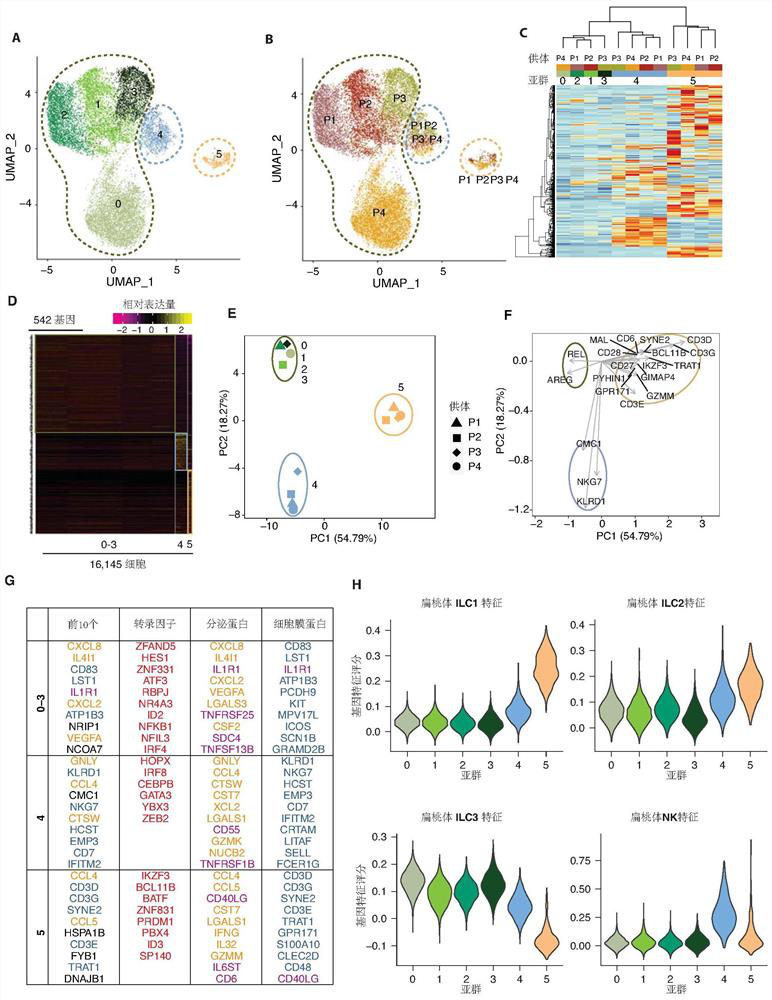
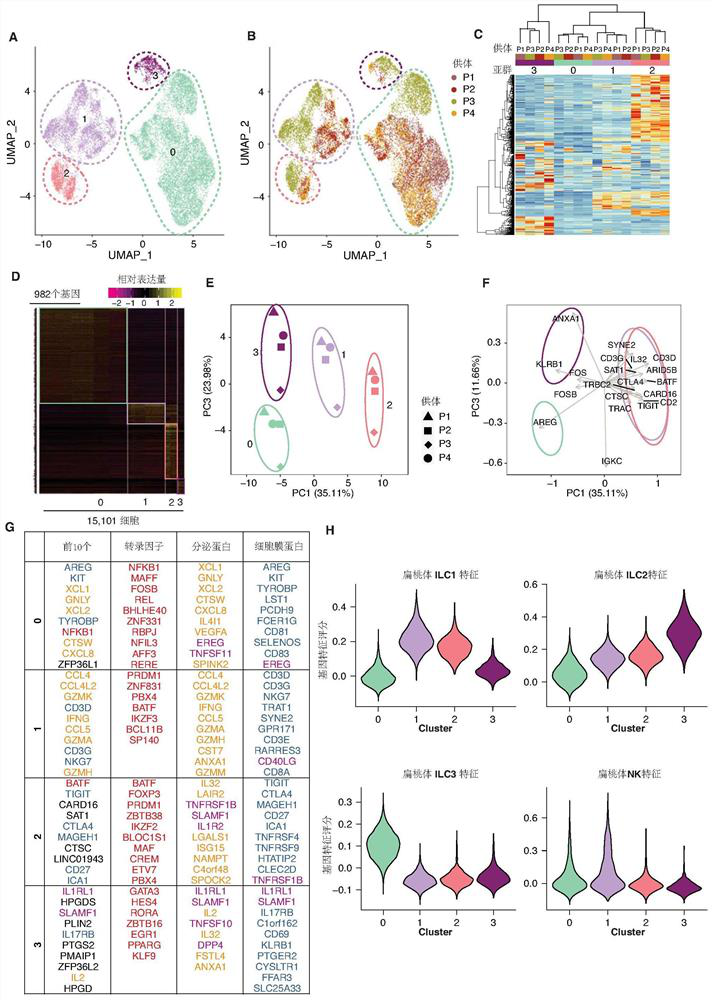
[0170] Example 3 Hierarchical Clustering
[0171] 1. Materials and methods
[0172] (1.1) Unsupervised Hierarchical Clustering
[0173] Calculate gene expression values for individual cells in each cell population. Only genes selected previously as variable features are used. The present invention uses the Heatmap.plus package to draw an unsupervised cluster map. Euclidean distances were calculated for genes in all cell populations. For normal mucosa, only cell populations clustered 0-4 were used for analysis.
[0174] (1.2) Dimensionality reduction and clustering
[0175] The top 2000 genes were screened using Seurat's "FindVariableGenes" function (Stuart et al., 2019) and used for principal component analysis (PCA). For ILCs in normal mucosa, the present invention retains the first 40 principal components. For normal blood, CRC blood and tumor tissue, the present invention retains the top 20 PCs. Clusters are identified using the "FindClusters" function, which is b...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com