Method for identifying three original species of rhei radix et rhizoma medicinal material and application
A technology of yellow medicinal materials and medicinal materials, which is applied in the field of identification of source varieties of Chinese medicinal materials, and can solve problems such as cumbersome operations and difficult origin species
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0021] Example 1 Screening of three primitive species-specific DNA barcodes of rhubarb medicinal materials
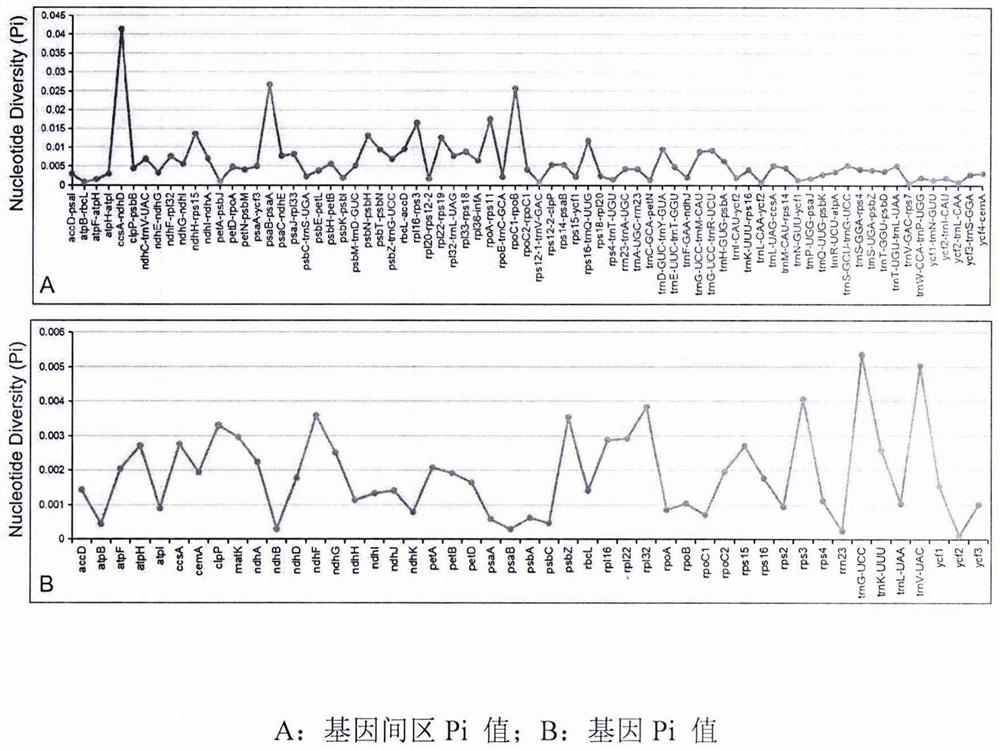
[0022] Taking the annotated chloroplast genome of Rhubarb palmatum as a reference, the chloroplast genomes of Rheum tangutica Rh. figure 1 ). The variation of the intergenic region of the chloroplast genome sequence of the three species is greater than that of the gene region, such as rps16-trnQ, trnT-psbD, psaA-ycf3, trnT-trnL, psbE-petL, ndhF-rpl32, trnN-ycf1, ccsA-ndhD, etc., these high The variable regions may be potential molecular markers for species identification of Rheum tangutica, Rheum officinalis and Rhubarb palmatum.
[0023] DnaSP was used to calculate the nucleotide variation value (Pi) of all genes and intergenic regions of the chloroplast genomes of the three species, and to perform hypervariable region detection and sequence difference level analysis ( figure 2 ). The variation of the mean value of intergenic region Pi (0.004) in the chloroplast ge...
Embodiment 2
[0024] Example 2 Acquisition of three primitive species-specific DNA barcodes of rhubarb medicinal materials
[0025] 2.1 Experimental materials
[0026] A total of 42 samples were collected in this example, including the three basic species of rhubarb medicinal materials, Rheumtanguticum, medicinal rhubarb Rh.officinale and palm leaf rhubarb Rh.palmatum, as well as edible rhubarb Rh.rhaponticum and wave leaf rhubarb Rh.rhabarbarum Linnaeus, Rh.alexandrae Batal., Rh.nobile Hook.f.et Thoms. and Rumex crispus. See Table 2 for details.
[0027] Table 2 Sample information table of the three basic species of rhubarb medicinal materials and their counterfeit products
[0028]
[0029] 2.2 Genome Extraction
[0030] About 50-100 mg of the samples in Table 2 were taken, ground with liquid nitrogen, and the rest of the steps were carried out according to the standard operating method of the plant genomic DNA extraction kit.
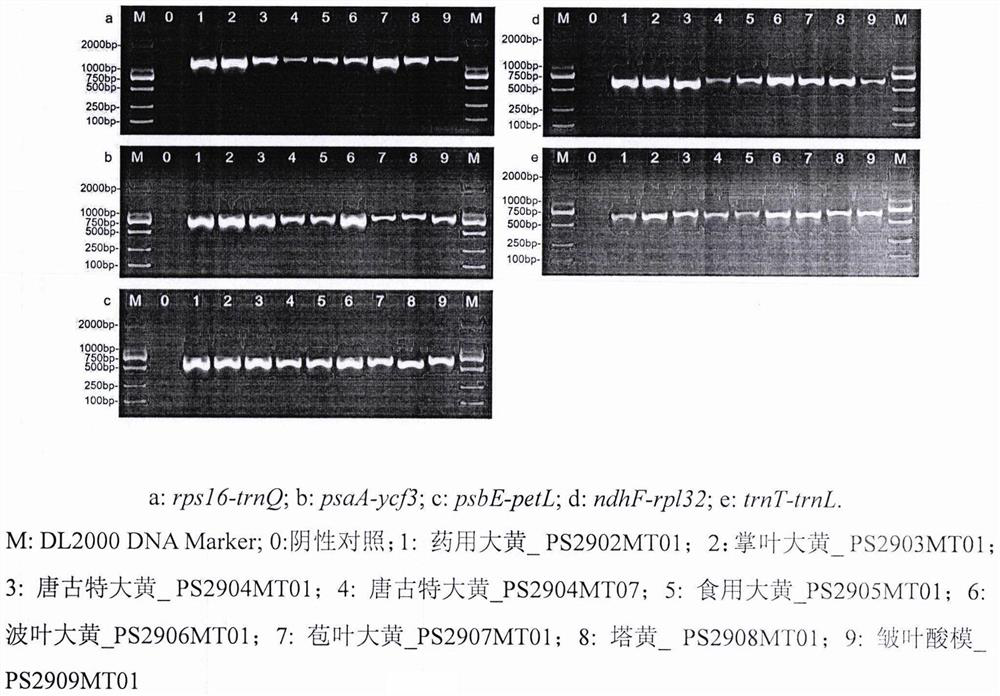
[0031] 2.3 PCR amplification
[0032] Primers 1F / 1R, ...
Embodiment 3
[0039] Example 3 Identification and analysis of three primitive species-specific DNA barcodes of rhubarb medicinal materials
[0040] 3.1 K2P distance
[0041] The intra-species and inter-species K2P distances of the high-quality sequences obtained by sequencing were calculated according to MEGA7.0. In the present invention, the minimum genetic distance between species is greater than the maximum genetic distance within a species, and there is no overlap, indicating that the obtained sequences meet the requirements of DNA barcodes (Table 3).
[0042] 3.2 Phylogenetic tree of NJ
[0043]The NJ phylogenetic tree was constructed according to MEGA7.0, and the repetition value of the bootstrap was set to 1000. In the present invention, rhubarb tangutica is gathered into one branch alone, rhubarb palmata is gathered into one branch alone, and rhubarb palmata is gathered into one branch alone, indicating that rpsl6-trnQ, psaA-ycf3, psbE-petL, ndhF-rpl32 and trnT -trnL can be used ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com