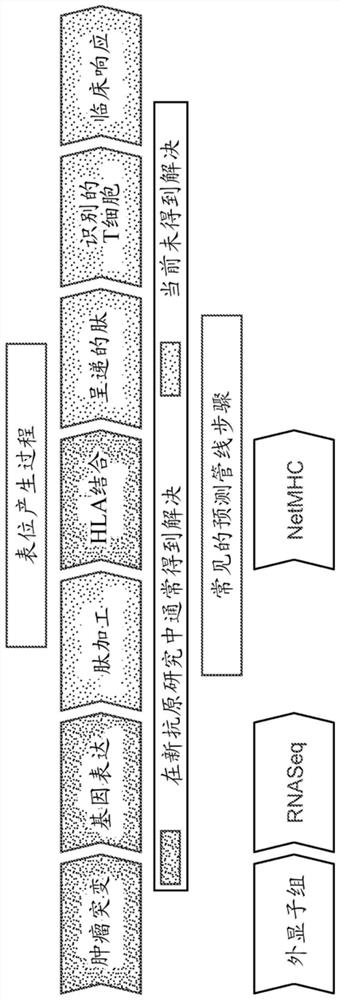
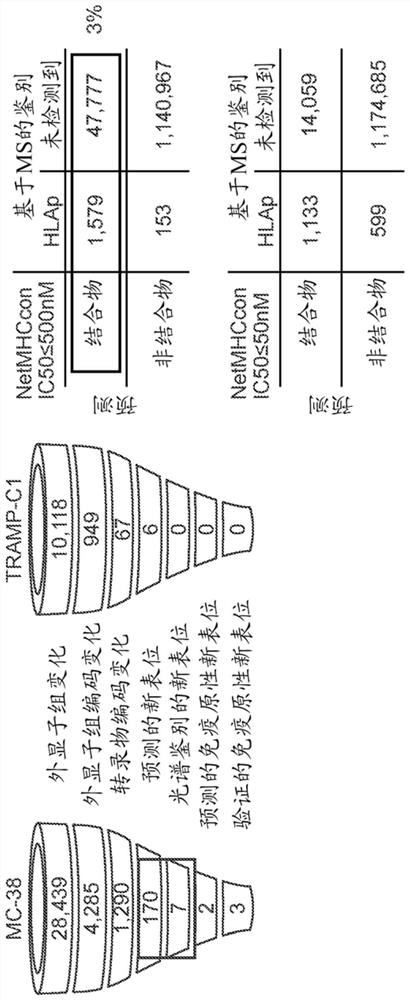
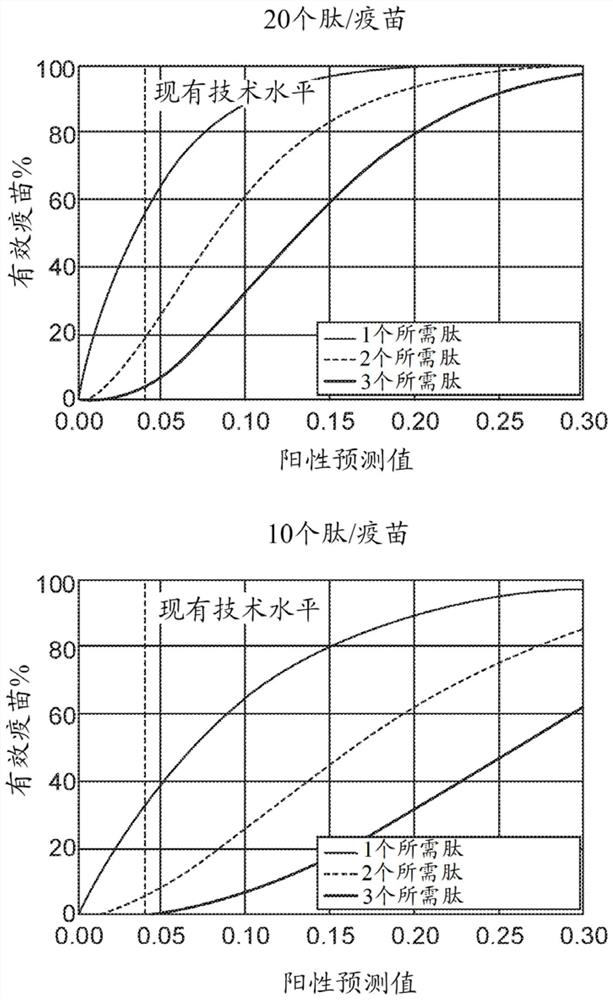
Identification of neo-antigens with mhc class ii model
A technology of antigens and models, applied in biological neural network models, antibody medical components, special data processing applications, etc., can solve the problems of missing candidate neoantigens, inefficient use of autoimmunity in vaccines, etc., and achieve the effect of speeding up the process
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example
[0423] In another embodiment, the deviation parameter θ h 0 May be shared by gene families of the MHC allele h. That is, the bias parameter θ of the MHC allele h h 0 can be equal to θ 基因(h) 0 , where gene (h) is the gene family of the MHC allele h. For example, the MHC class I alleles HLA-A*02:01, HLA-A*02:02, and HLA-A*02:03 can be assigned to the "HLA-A" gene family, and these MHC alleles The respective bias parameters θ h 0 Can be shared. As another example, the MHC class II alleles HLA-DRB1:10:01, HLA-DRB1:11:01, and HLA-DRB3:01:01 may be assigned to the "HLA-DRB" gene family, and these MHC alleles Gene's respective bias parameter θ h 0 Can be shared.
[0424] Going back to equation (2), as an example, when using the affine correlation function g h (·) Of the m = 4 different MHC alleles identified, peptide p k The probability of being presented by the MHC allele h=3 can be given by:
[0425]
[0426] where x 3 k is the allelic interaction variable for t...
Embodiment 1
[0470] VIII.C.1. Example 1: Maximum of Independent Allele Models
[0471] In one embodiment, the training module 316 causes peptide p associated with a set of multiple MHC alleles H k The estimated probability of presentation u k With the probability of presentation of each MHC allele h in set H determined based on cells expressing the monoallele The variation of is modeled as described above in connection with equations (2)-(11). Specifically, the presentation probability u k can be any function of . In one embodiment, as shown in equation (12), this function is a maximum function and renders the likelihood u k can be determined as the maximum probability of presentation for each MHC allele h in set H.
[0472]
[0473] VIII.C.2. Example 2.1: The Funciton-of-Sums Model
[0474] In one embodiment, the training module 316 makes peptide p by k The estimated probability of presentation u k Modeling:
[0475]
[0476] where element a h k For the peptide seq...
Embodiment 22
[0486] VIII.C.3. Example 2.2: Functional Model Using the Sum of Allelic Non-Interacting Variables
[0487] In one embodiment, the training module 316 incorporates allelic non-interaction variables and makes peptide p by k The estimated probability of presentation u k Modeling:
[0488]
[0489] where w k Indicates the encoded related peptide p k The allelic non-interaction variable for . Specifically, the parameter set θ for each MHC allele h h and the set of parameters about the allelic non-interacting variables θ w The value of θ can be obtained by making about θ h and θ w is determined by minimizing a loss function of , where i is each instance in the subset S of training data 170 produced by cells expressing a single MHC allele and / or by cells expressing multiple MHC alleles. Correlation function g w The correlation function g introduced in Section VIII.B.3 above can be expressed as w any of the forms.
[0490] Therefore, according to equation (14), the fun...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com