Construction method of plant paired NLR resistance gene database and multi-species paired NLR gene database thereof
A resistance gene and construction method technology, applied in the field of gene sequence data processing, can solve the problems of plant disease resistance gene cloning, analysis and application obstacles, frequent NLR gene duplication and loss, uneven genome distribution, etc., to achieve accelerated resistance The effect of disease breeding
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
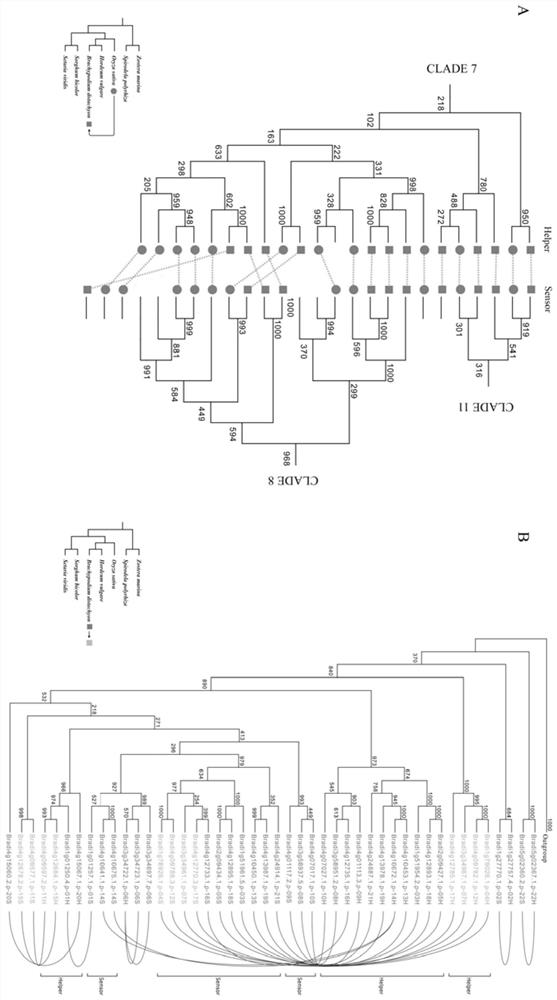
[0044] Identification of paired NLR genes in Brachypodium distachyon
[0045] 1. Collection of basic data: Collect the genome sequence, protein sequence and gene annotation information of Brachypodium distachyon from the JGI public database (https: / / phytozome.jgi.doe.gov);
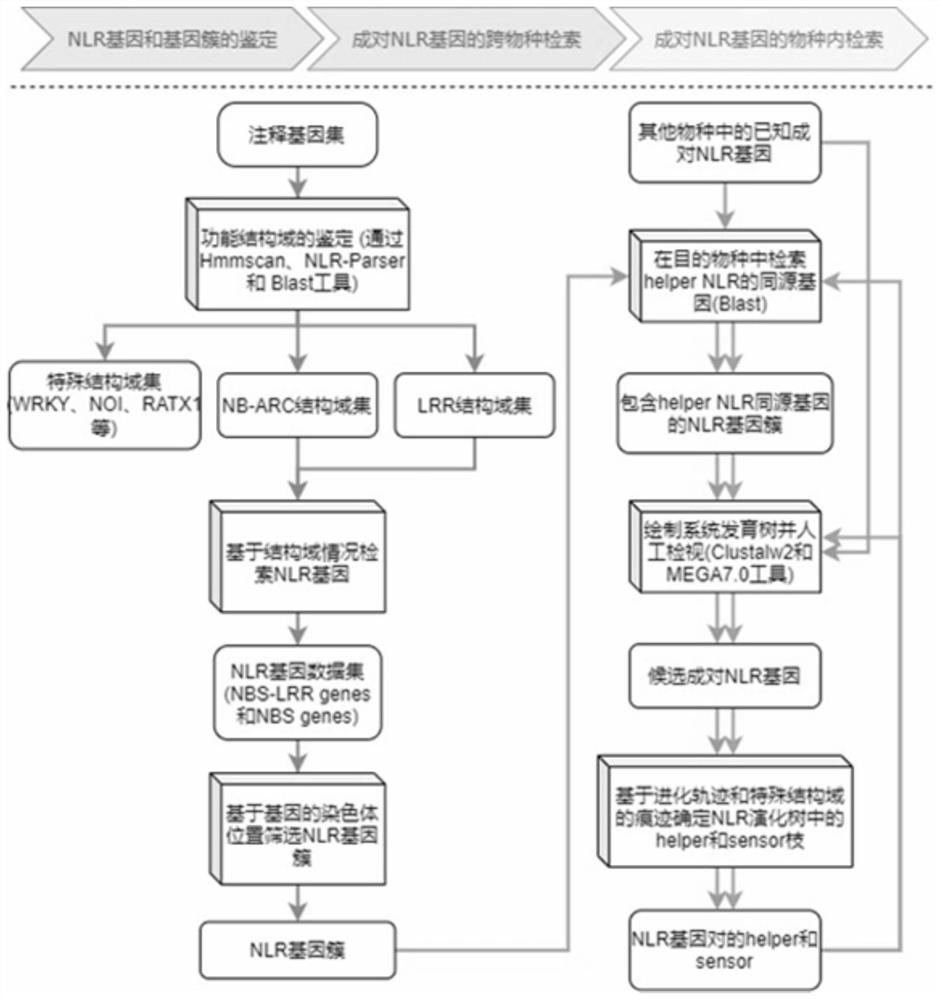
[0046] 2. Identification of NLR genes and gene clusters: (1) Firstly, based on the protein sequence information of the whole genome, obtain the structural domain information of all protein sequences through the Hmmscan tool, and screen the proteins containing the NB-ARC domain (PF00931), parameters Set to E-value≤1e-4; (2) Then use the NLR-parser tool to detect the LRR domain and screen for proteins with motif 9, 11 or 19, and have both the NB-ARC domain and the LRR domain The gene of the NLR gene is the main NLR gene, and the gene with only the NB-ARC domain is the candidate NLR gene; (3) Then based on the physical distance and gene number of the NLR gene on the chromosome in the gene annotation, we put t...
Embodiment 2
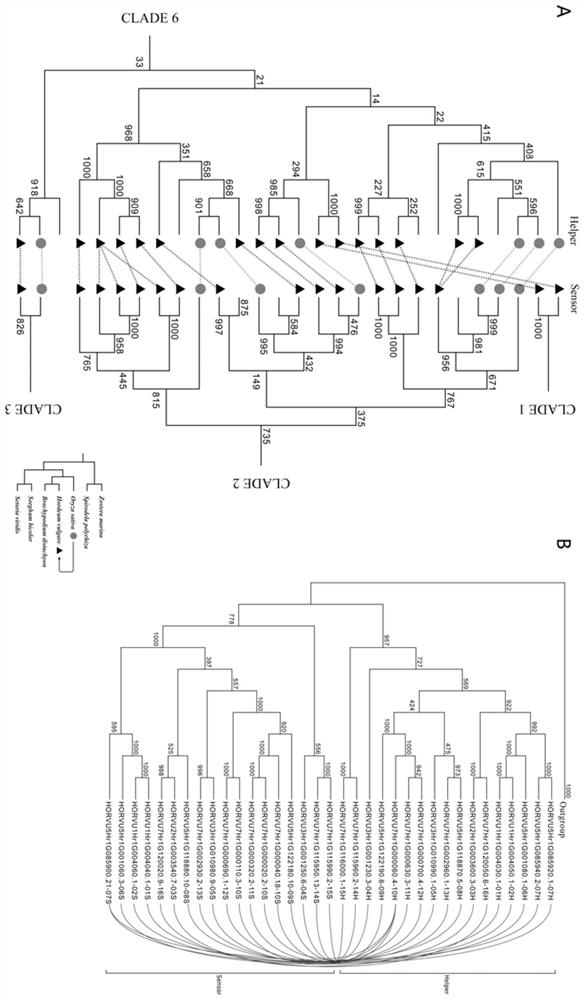
[0052] Identification of paired NLR genes in barley (Hordeum vulgare)
[0053]1. Collection of basic data: Collect genome sequence, protein sequence and gene annotation information of barley from Ensembl public database (http: / / plants.ensembl.org);
[0054] 2. Identification of NLR genes and gene clusters: (1) Firstly, based on the protein sequence information of the whole genome, obtain the structural domain information of all protein sequences through the Hmmscan tool, and screen the proteins containing the NB-ARC domain (PF00931), parameters Set to E-value≤1e-4; (2) Then use the NLR-parser tool to detect the LRR domain and screen for proteins with motif 9, 11 or 19, and have both the NB-ARC domain and the LRR domain The gene of the NLR gene is the main NLR gene, and the gene with only the NB-ARC domain is the candidate NLR gene; (3) Then based on the physical distance and gene number of the NLR gene on the chromosome in the gene annotation, we put the physical distance on t...
Embodiment 3
[0060] Identification of paired NLR genes in sorghum (Sorghum bicolor)
[0061] 1. Collection of basic data: Collect genome sequence, protein sequence and gene annotation information of sorghum from JGI public database (https: / / phytozome.jgi.doe.gov);
[0062] 2. Identification of NLR genes and gene clusters: (1) Firstly, based on the protein sequence information of the whole genome, obtain the structural domain information of all protein sequences through the Hmmscan tool, and screen the proteins containing the NB-ARC domain (PF00931), parameters Set to E-value≤1e-4; (2) Then use the NLR-parser tool to detect the LRR domain and screen for proteins with motif 9, 11 or 19, and have both the NB-ARC domain and the LRR domain The gene of the NLR gene is the main NLR gene, and the gene with only the NB-ARC domain is the candidate NLR gene; (3) Then based on the physical distance and gene number of the NLR gene on the chromosome in the gene annotation, we put the physical distance o...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com